Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6909
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7489 | 3.843e-03 | 6.410e-04 | 7.829e-01 | 1.929e-06 |
|---|
| Loi | 0.2306 | 7.974e-02 | 1.122e-02 | 4.269e-01 | 3.819e-04 |
|---|
| Schmidt | 0.6764 | 0.000e+00 | 0.000e+00 | 4.184e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7911 | 0.000e+00 | 0.000e+00 | 3.429e-03 | 0.000e+00 |
|---|
| Wang | 0.2638 | 2.397e-03 | 4.118e-02 | 3.838e-01 | 3.789e-05 |
|---|
Expression data for subnetwork 6909 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
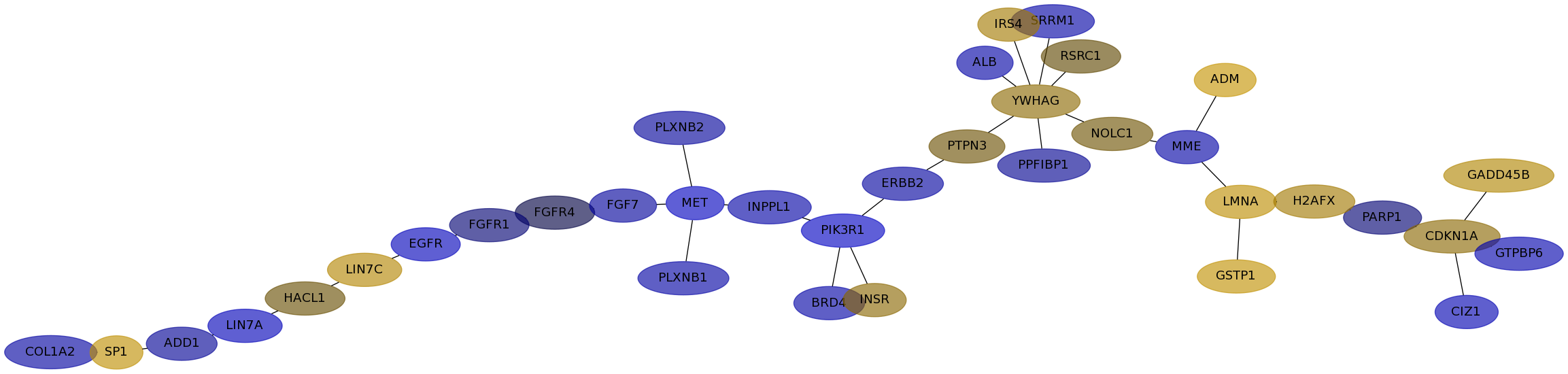
Score for each gene in subnetwork 6909 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| HACL1 |   | 4 | 440 | 590 | 585 | 0.028 | -0.071 | undef | -0.278 | undef |
|---|
| INPPL1 |   | 6 | 301 | 141 | 152 | -0.122 | 0.072 | -0.087 | 0.174 | 0.156 |
|---|
| ADD1 |   | 8 | 222 | 366 | 373 | -0.085 | 0.116 | 0.035 | undef | -0.098 |
|---|
| SRRM1 |   | 10 | 167 | 296 | 283 | -0.120 | 0.139 | 0.244 | 0.294 | -0.109 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| PTPN3 |   | 11 | 148 | 658 | 635 | 0.029 | 0.060 | -0.067 | 0.225 | -0.034 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| GSTP1 |   | 14 | 117 | 1 | 34 | 0.222 | -0.166 | 0.201 | 0.126 | -0.136 |
|---|
| RSRC1 |   | 21 | 73 | 366 | 354 | 0.023 | 0.104 | 0.105 | -0.098 | 0.212 |
|---|
| LMNA |   | 3 | 557 | 1047 | 1038 | 0.208 | 0.223 | -0.017 | 0.247 | 0.069 |
|---|
| PLXNB1 |   | 4 | 440 | 1047 | 1032 | -0.117 | 0.312 | 0.006 | -0.172 | -0.146 |
|---|
| PPFIBP1 |   | 2 | 743 | 1047 | 1055 | -0.073 | 0.207 | -0.072 | 0.085 | -0.016 |
|---|
| LIN7A |   | 6 | 301 | 366 | 377 | -0.189 | -0.074 | 0.108 | 0.065 | -0.146 |
|---|
| CIZ1 |   | 2 | 743 | 1047 | 1055 | -0.162 | 0.045 | -0.006 | 0.139 | -0.108 |
|---|
| MET |   | 6 | 301 | 1047 | 1020 | -0.219 | 0.086 | 0.183 | -0.025 | 0.170 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| FGFR4 |   | 21 | 73 | 179 | 183 | -0.011 | 0.188 | -0.089 | -0.018 | -0.049 |
|---|
| BRD4 |   | 11 | 148 | 1047 | 1003 | -0.119 | 0.095 | -0.075 | 0.081 | -0.044 |
|---|
| PARP1 |   | 29 | 45 | 83 | 84 | -0.036 | -0.162 | -0.089 | 0.238 | 0.012 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| LIN7C |   | 10 | 167 | 590 | 566 | 0.166 | 0.088 | 0.118 | -0.018 | 0.094 |
|---|
| IRS4 |   | 32 | 40 | 236 | 224 | 0.115 | -0.111 | -0.088 | 0.096 | 0.142 |
|---|
| PLXNB2 |   | 4 | 440 | 590 | 585 | -0.092 | 0.308 | -0.136 | undef | -0.062 |
|---|
| ADM |   | 10 | 167 | 682 | 653 | 0.255 | 0.187 | 0.091 | 0.169 | 0.212 |
|---|
| FGFR1 |   | 26 | 52 | 638 | 615 | -0.036 | 0.200 | 0.087 | undef | -0.015 |
|---|
| YWHAG |   | 24 | 62 | 296 | 279 | 0.067 | 0.024 | undef | 0.266 | undef |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| GTPBP6 |   | 6 | 301 | 928 | 907 | -0.143 | -0.131 | -0.167 | 0.155 | -0.129 |
|---|
| ALB |   | 3 | 557 | 412 | 430 | -0.122 | 0.218 | 0.080 | 0.018 | 0.038 |
|---|
| GADD45B |   | 8 | 222 | 318 | 325 | 0.158 | 0.122 | 0.067 | 0.060 | 0.212 |
|---|
| H2AFX |   | 7 | 256 | 179 | 201 | 0.111 | -0.072 | 0.121 | 0.165 | -0.010 |
|---|
| FGF7 |   | 10 | 167 | 780 | 760 | -0.094 | 0.237 | 0.070 | 0.038 | 0.083 |
|---|
| COL1A2 |   | 7 | 256 | 552 | 542 | -0.110 | 0.207 | 0.046 | 0.235 | 0.097 |
|---|
| NOLC1 |   | 4 | 440 | 1047 | 1032 | 0.033 | -0.055 | 0.172 | 0.012 | -0.064 |
|---|
| MME |   | 5 | 360 | 1047 | 1021 | -0.121 | 0.020 | 0.088 | -0.002 | -0.130 |
|---|
GO Enrichment output for subnetwork 6909 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.885E-08 | 4.606E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.234E-06 | 1.508E-03 |
|---|
| positive regulation of axonogenesis | GO:0050772 |  | 9.884E-06 | 8.049E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.762E-05 | 0.01075903 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.854E-05 | 0.01394223 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 3.423E-05 | 0.01393643 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 4.316E-05 | 0.01506163 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 4.607E-05 | 0.01406764 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 4.875E-05 | 0.0132336 |
|---|
| interaction with host | GO:0051701 |  | 4.894E-05 | 0.01195612 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 6.065E-05 | 0.01346986 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 9.397E-07 | 2.261E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.478E-06 | 1.778E-03 |
|---|
| positive regulation of axonogenesis | GO:0050772 |  | 1.183E-05 | 9.485E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.76E-05 | 0.01058514 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.931E-05 | 0.01410358 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 3.341E-05 | 0.01339883 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 3.942E-05 | 0.01354925 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 5.168E-05 | 0.01554279 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 5.478E-05 | 0.01464553 |
|---|
| interaction with host | GO:0051701 |  | 5.849E-05 | 0.01407242 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 6.597E-05 | 0.01443016 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.496E-06 | 5.74E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 3.543E-06 | 4.074E-03 |
|---|
| positive regulation of axonogenesis | GO:0050772 |  | 2.263E-05 | 0.01734865 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 3.281E-05 | 0.01886854 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 4.191E-05 | 0.01927633 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 5.937E-05 | 0.02275755 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 6.964E-05 | 0.02288199 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 8.101E-05 | 0.02329071 |
|---|
| interaction with host | GO:0051701 |  | 1.072E-04 | 0.0274073 |
|---|
| positive regulation of cell projection organization | GO:0031346 |  | 1.222E-04 | 0.02810836 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.222E-04 | 0.02555305 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 6.405E-06 | 0.01180384 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.334E-05 | 0.01229311 |
|---|
| interaction with host | GO:0051701 |  | 1.334E-05 | 8.195E-03 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.773E-05 | 8.17E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.298E-05 | 8.471E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 2.916E-05 | 8.957E-03 |
|---|
| positive regulation of protein kinase activity | GO:0045860 |  | 3.571E-05 | 9.403E-03 |
|---|
| positive regulation of transferase activity | GO:0051347 |  | 3.992E-05 | 9.196E-03 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 6.204E-05 | 0.01270446 |
|---|
| symbiosis. encompassing mutualism through parasitism | GO:0044403 |  | 6.458E-05 | 0.01190172 |
|---|
| positive regulation of glucose metabolic process | GO:0010907 |  | 1.949E-04 | 0.0326464 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.496E-06 | 5.74E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 3.543E-06 | 4.074E-03 |
|---|
| positive regulation of axonogenesis | GO:0050772 |  | 2.263E-05 | 0.01734865 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 3.281E-05 | 0.01886854 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 4.191E-05 | 0.01927633 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 5.937E-05 | 0.02275755 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 6.964E-05 | 0.02288199 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 8.101E-05 | 0.02329071 |
|---|
| interaction with host | GO:0051701 |  | 1.072E-04 | 0.0274073 |
|---|
| positive regulation of cell projection organization | GO:0031346 |  | 1.222E-04 | 0.02810836 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.222E-04 | 0.02555305 |
|---|



