Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6902
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7490 | 3.835e-03 | 6.400e-04 | 7.827e-01 | 1.921e-06 |
|---|
| Loi | 0.2306 | 7.974e-02 | 1.122e-02 | 4.269e-01 | 3.821e-04 |
|---|
| Schmidt | 0.6763 | 0.000e+00 | 0.000e+00 | 4.188e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7911 | 0.000e+00 | 0.000e+00 | 3.427e-03 | 0.000e+00 |
|---|
| Wang | 0.2637 | 2.410e-03 | 4.131e-02 | 3.844e-01 | 3.827e-05 |
|---|
Expression data for subnetwork 6902 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
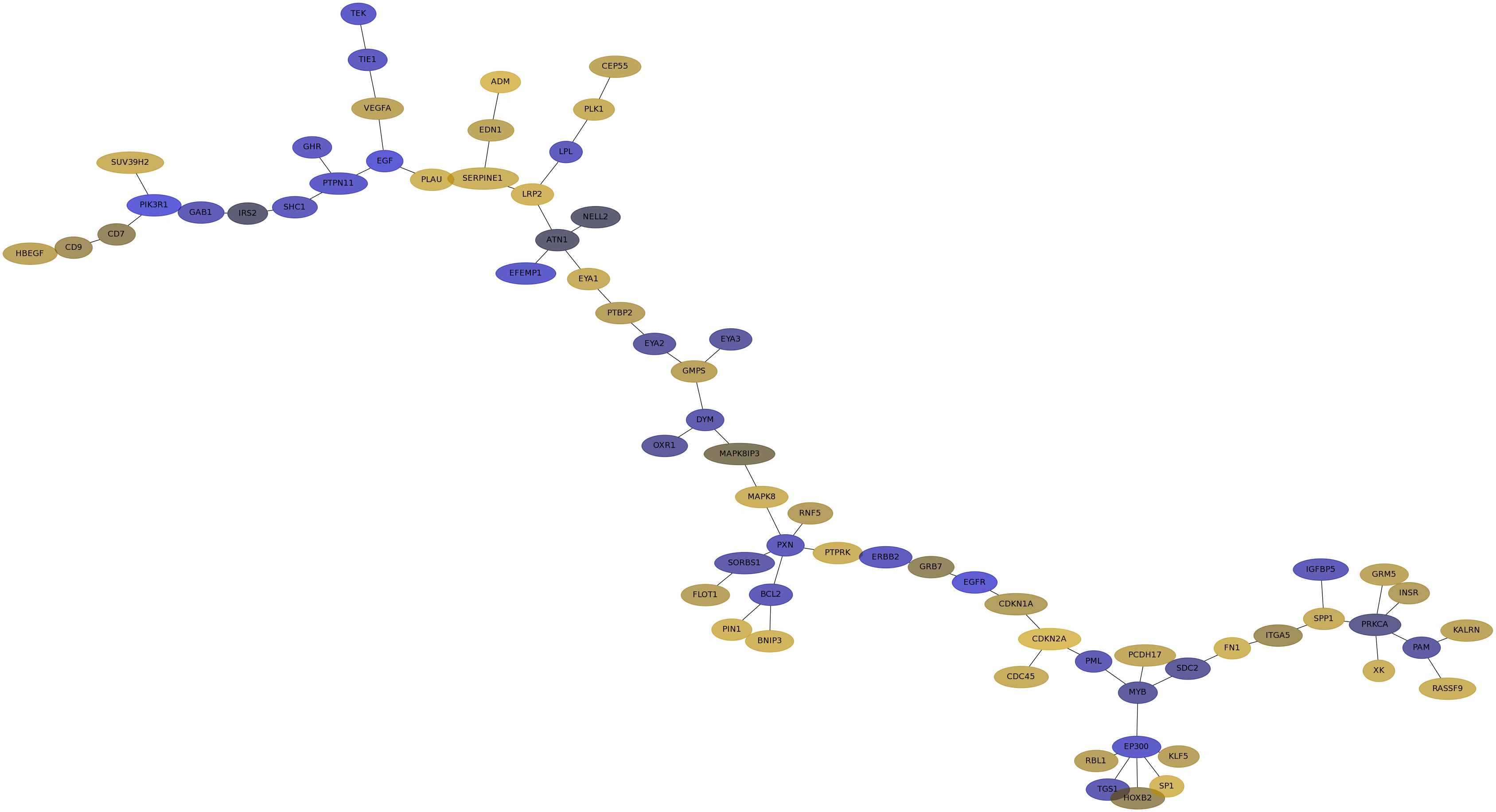
Score for each gene in subnetwork 6902 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| MAPK8 |   | 8 | 222 | 696 | 677 | 0.153 | 0.200 | 0.249 | -0.066 | 0.138 |
|---|
| TEK |   | 18 | 86 | 83 | 95 | -0.149 | -0.049 | 0.174 | 0.015 | 0.113 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| HBEGF |   | 1 | 1195 | 1102 | 1139 | 0.087 | 0.002 | 0.039 | 0.036 | 0.076 |
|---|
| PCDH17 |   | 1 | 1195 | 1102 | 1139 | 0.109 | -0.064 | undef | 0.135 | undef |
|---|
| EGF |   | 12 | 140 | 638 | 620 | -0.212 | 0.182 | -0.276 | -0.060 | 0.005 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| GRM5 |   | 15 | 111 | 179 | 191 | 0.094 | -0.019 | 0.000 | 0.093 | -0.093 |
|---|
| NELL2 |   | 7 | 256 | 727 | 714 | -0.005 | 0.138 | 0.421 | -0.027 | 0.074 |
|---|
| SHC1 |   | 4 | 440 | 478 | 479 | -0.090 | -0.041 | -0.005 | 0.169 | -0.138 |
|---|
| PLAU |   | 12 | 140 | 179 | 192 | 0.187 | 0.153 | 0.141 | 0.072 | 0.192 |
|---|
| PIN1 |   | 24 | 62 | 179 | 181 | 0.194 | 0.040 | -0.040 | 0.065 | -0.048 |
|---|
| PTPRK |   | 14 | 117 | 83 | 96 | 0.156 | 0.141 | -0.094 | 0.048 | -0.024 |
|---|
| PLK1 |   | 11 | 148 | 236 | 232 | 0.142 | -0.143 | 0.094 | -0.192 | -0.163 |
|---|
| PTBP2 |   | 2 | 743 | 1102 | 1102 | 0.073 | 0.057 | 0.131 | -0.114 | 0.117 |
|---|
| EYA3 |   | 9 | 196 | 318 | 324 | -0.034 | 0.053 | -0.020 | 0.188 | -0.142 |
|---|
| KALRN |   | 4 | 440 | 1102 | 1080 | 0.090 | 0.136 | -0.057 | 0.202 | -0.053 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| ADM |   | 10 | 167 | 682 | 653 | 0.255 | 0.187 | 0.091 | 0.169 | 0.212 |
|---|
| CDC45 |   | 5 | 360 | 318 | 334 | 0.134 | -0.171 | 0.185 | -0.103 | -0.085 |
|---|
| IRS2 |   | 3 | 557 | 1102 | 1098 | -0.006 | 0.043 | 0.054 | -0.256 | 0.125 |
|---|
| RBL1 |   | 21 | 73 | 1 | 27 | 0.078 | undef | undef | 0.051 | undef |
|---|
| CEP55 |   | 2 | 743 | 928 | 929 | 0.097 | -0.196 | 0.056 | -0.002 | 0.052 |
|---|
| PAM |   | 10 | 167 | 638 | 621 | -0.035 | 0.307 | 0.200 | 0.002 | 0.104 |
|---|
| LPL |   | 13 | 124 | 236 | 230 | -0.102 | -0.062 | 0.292 | 0.087 | -0.105 |
|---|
| SERPINE1 |   | 8 | 222 | 179 | 200 | 0.160 | 0.200 | 0.056 | 0.070 | 0.030 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| TGS1 |   | 5 | 360 | 780 | 767 | -0.060 | -0.044 | 0.232 | undef | 0.269 |
|---|
| GAB1 |   | 5 | 360 | 1102 | 1076 | -0.069 | 0.084 | -0.117 | 0.116 | -0.212 |
|---|
| EYA2 |   | 13 | 124 | 318 | 316 | -0.034 | 0.113 | -0.142 | undef | -0.019 |
|---|
| FN1 |   | 6 | 301 | 590 | 582 | 0.190 | 0.133 | 0.091 | undef | 0.083 |
|---|
| RASSF9 |   | 4 | 440 | 1102 | 1080 | 0.156 | -0.058 | 0.173 | 0.122 | -0.133 |
|---|
| EYA1 |   | 8 | 222 | 1010 | 981 | 0.135 | -0.065 | 0.162 | 0.068 | -0.088 |
|---|
| DYM |   | 2 | 743 | 1102 | 1102 | -0.053 | -0.105 | -0.076 | 0.033 | 0.109 |
|---|
| SDC2 |   | 4 | 440 | 590 | 585 | -0.027 | 0.068 | 0.033 | undef | 0.215 |
|---|
| MAPK8IP3 |   | 3 | 557 | 928 | 925 | 0.010 | 0.045 | -0.046 | undef | 0.059 |
|---|
| LRP2 |   | 3 | 557 | 1102 | 1098 | 0.193 | -0.042 | 0.001 | -0.138 | 0.125 |
|---|
| FLOT1 |   | 2 | 743 | 1102 | 1102 | 0.076 | 0.004 | -0.095 | 0.188 | -0.095 |
|---|
| CD7 |   | 2 | 743 | 1102 | 1102 | 0.020 | -0.151 | -0.165 | 0.027 | -0.073 |
|---|
| HOXB2 |   | 33 | 37 | 318 | 289 | 0.024 | -0.143 | 0.076 | 0.247 | -0.184 |
|---|
| EFEMP1 |   | 1 | 1195 | 1102 | 1139 | -0.148 | 0.011 | 0.146 | -0.201 | 0.067 |
|---|
| SPP1 |   | 26 | 52 | 83 | 91 | 0.130 | 0.119 | 0.184 | 0.204 | 0.153 |
|---|
| SUV39H2 |   | 8 | 222 | 318 | 325 | 0.150 | -0.097 | -0.057 | undef | 0.028 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| PTPN11 |   | 20 | 81 | 83 | 94 | -0.152 | 0.014 | 0.066 | 0.304 | 0.089 |
|---|
| CD9 |   | 13 | 124 | 525 | 503 | 0.036 | -0.115 | -0.109 | 0.281 | -0.147 |
|---|
| GMPS |   | 7 | 256 | 318 | 328 | 0.083 | -0.022 | 0.044 | -0.019 | 0.151 |
|---|
| BNIP3 |   | 11 | 148 | 236 | 232 | 0.193 | 0.071 | 0.102 | 0.119 | 0.285 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| RNF5 |   | 2 | 743 | 1102 | 1102 | 0.067 | 0.024 | 0.050 | 0.035 | 0.252 |
|---|
| MYB |   | 28 | 47 | 366 | 344 | -0.028 | 0.075 | 0.002 | 0.065 | -0.012 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| PXN |   | 12 | 140 | 1 | 36 | -0.088 | 0.284 | -0.022 | 0.060 | 0.114 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| OXR1 |   | 1 | 1195 | 1102 | 1139 | -0.029 | -0.183 | undef | undef | undef |
|---|
| EDN1 |   | 6 | 301 | 682 | 669 | 0.100 | 0.126 | 0.249 | -0.086 | 0.121 |
|---|
| ITGA5 |   | 26 | 52 | 179 | 177 | 0.031 | 0.192 | 0.125 | 0.133 | 0.202 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| XK |   | 18 | 86 | 179 | 187 | 0.154 | -0.000 | 0.067 | -0.123 | -0.040 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| PML |   | 6 | 301 | 478 | 473 | -0.068 | -0.010 | -0.007 | -0.153 | -0.070 |
|---|
| ATN1 |   | 7 | 256 | 412 | 414 | -0.005 | -0.093 | 0.005 | -0.185 | -0.136 |
|---|
| TIE1 |   | 18 | 86 | 366 | 357 | -0.108 | 0.005 | 0.141 | -0.108 | 0.252 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
GO Enrichment output for subnetwork 6902 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 8.591E-13 | 2.099E-09 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.315E-12 | 1.606E-09 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 2.368E-12 | 1.928E-09 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 7.508E-11 | 4.585E-08 |
|---|
| organ growth | GO:0035265 |  | 7.534E-11 | 3.681E-08 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 7.1E-10 | 2.891E-07 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.017E-09 | 3.548E-07 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 1.596E-09 | 4.875E-07 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.812E-09 | 4.92E-07 |
|---|
| activation of MAPK activity | GO:0000187 |  | 2.321E-09 | 5.671E-07 |
|---|
| response to insulin stimulus | GO:0032868 |  | 4.87E-09 | 1.082E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 4.214E-12 | 1.014E-08 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 2.994E-11 | 3.602E-08 |
|---|
| organ growth | GO:0035265 |  | 9.967E-11 | 7.994E-08 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.101E-10 | 6.625E-08 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 9.442E-10 | 4.544E-07 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 2.448E-09 | 9.816E-07 |
|---|
| activation of MAPK activity | GO:0000187 |  | 3.557E-09 | 1.223E-06 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 5.381E-09 | 1.618E-06 |
|---|
| response to hypoxia | GO:0001666 |  | 1.643E-08 | 4.393E-06 |
|---|
| response to oxygen levels | GO:0070482 |  | 1.996E-08 | 4.803E-06 |
|---|
| negative regulation of transport | GO:0051051 |  | 2.196E-08 | 4.804E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.996E-11 | 4.59E-08 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.093E-10 | 1.257E-07 |
|---|
| organ growth | GO:0035265 |  | 3.437E-10 | 2.635E-07 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 4.478E-10 | 2.575E-07 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 2.516E-09 | 1.157E-06 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 5.45E-09 | 2.089E-06 |
|---|
| activation of MAPK activity | GO:0000187 |  | 8.358E-09 | 2.746E-06 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 1.199E-08 | 3.447E-06 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.367E-08 | 3.493E-06 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 3.577E-08 | 8.228E-06 |
|---|
| response to hypoxia | GO:0001666 |  | 4.178E-08 | 8.735E-06 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cell migration | GO:0030334 |  | 2.722E-11 | 5.017E-08 |
|---|
| regulation of locomotion | GO:0040012 |  | 8.164E-11 | 7.523E-08 |
|---|
| regulation of cell motion | GO:0051270 |  | 8.164E-11 | 5.016E-08 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 9.292E-10 | 4.281E-07 |
|---|
| response to hypoxia | GO:0001666 |  | 1.79E-09 | 6.596E-07 |
|---|
| response to oxygen levels | GO:0070482 |  | 2.09E-09 | 6.42E-07 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 3.137E-09 | 8.258E-07 |
|---|
| regulation of anatomical structure morphogenesis | GO:0022603 |  | 4.502E-09 | 1.037E-06 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 5.691E-09 | 1.165E-06 |
|---|
| regulation of MAP kinase activity | GO:0043405 |  | 1.086E-08 | 2.001E-06 |
|---|
| negative regulation of multicellular organismal process | GO:0051241 |  | 6.069E-08 | 1.017E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.996E-11 | 4.59E-08 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.093E-10 | 1.257E-07 |
|---|
| organ growth | GO:0035265 |  | 3.437E-10 | 2.635E-07 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 4.478E-10 | 2.575E-07 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 2.516E-09 | 1.157E-06 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 5.45E-09 | 2.089E-06 |
|---|
| activation of MAPK activity | GO:0000187 |  | 8.358E-09 | 2.746E-06 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 1.199E-08 | 3.447E-06 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.367E-08 | 3.493E-06 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 3.577E-08 | 8.228E-06 |
|---|
| response to hypoxia | GO:0001666 |  | 4.178E-08 | 8.735E-06 |
|---|



