Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6885
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7490 | 3.835e-03 | 6.400e-04 | 7.826e-01 | 1.921e-06 |
|---|
| Loi | 0.2310 | 7.926e-02 | 1.110e-02 | 4.258e-01 | 3.745e-04 |
|---|
| Schmidt | 0.6760 | 0.000e+00 | 0.000e+00 | 4.221e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7911 | 0.000e+00 | 0.000e+00 | 3.425e-03 | 0.000e+00 |
|---|
| Wang | 0.2636 | 2.414e-03 | 4.135e-02 | 3.846e-01 | 3.840e-05 |
|---|
Expression data for subnetwork 6885 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
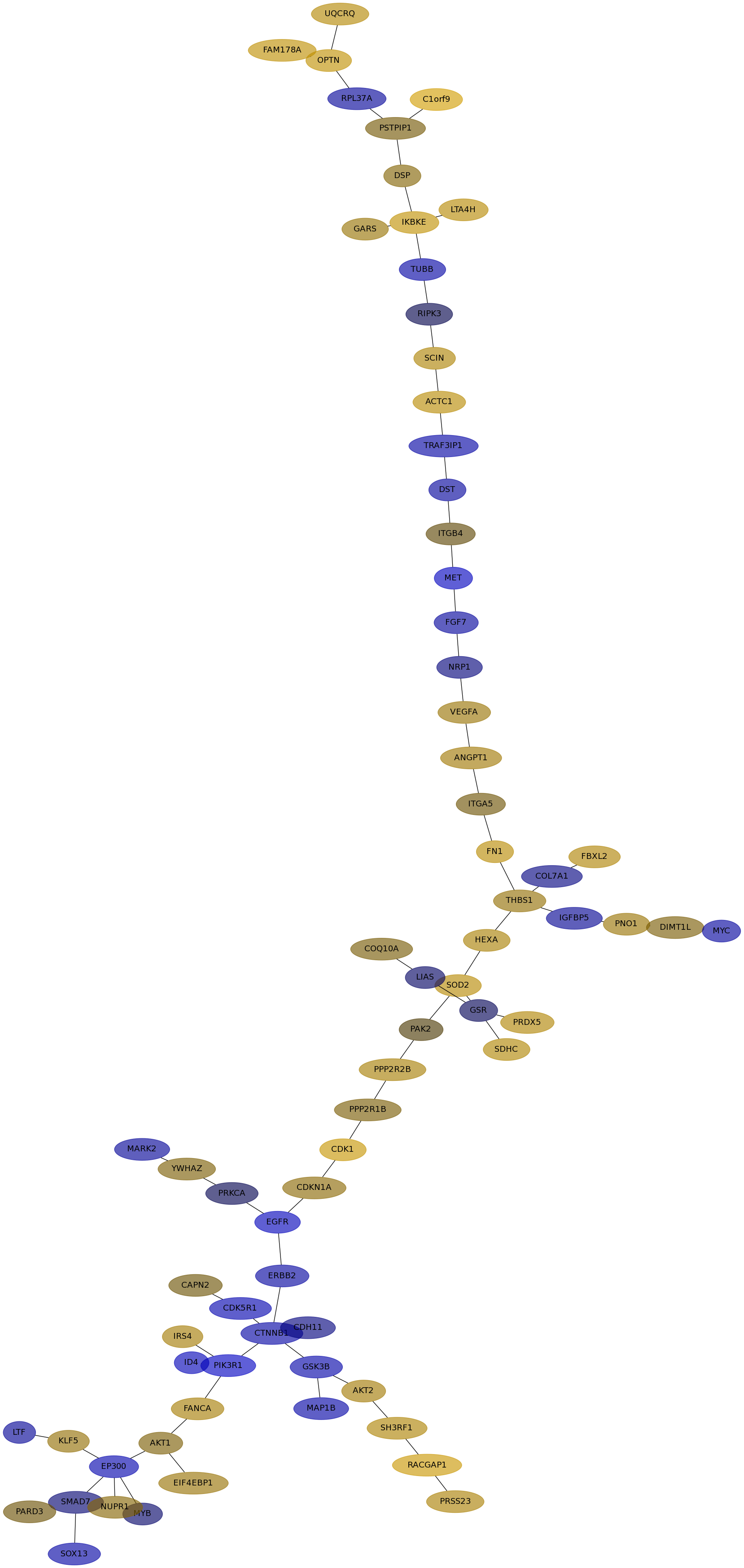
Score for each gene in subnetwork 6885 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| SOD2 |   | 1 | 1195 | 1222 | 1242 | 0.176 | -0.056 | -0.051 | -0.159 | -0.019 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| ID4 |   | 35 | 34 | 236 | 223 | -0.179 | 0.137 | 0.241 | undef | 0.036 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| LTF |   | 3 | 557 | 1222 | 1210 | -0.082 | 0.060 | -0.268 | -0.081 | -0.280 |
|---|
| PPP2R1B |   | 6 | 301 | 141 | 152 | 0.042 | 0.002 | -0.002 | undef | 0.112 |
|---|
| SCIN |   | 2 | 743 | 236 | 272 | 0.143 | -0.065 | 0.110 | undef | 0.132 |
|---|
| DST |   | 22 | 69 | 1 | 25 | -0.094 | 0.343 | 0.065 | -0.062 | 0.194 |
|---|
| GSR |   | 3 | 557 | 1222 | 1210 | -0.018 | -0.011 | -0.194 | 0.202 | -0.126 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| RPL37A |   | 1 | 1195 | 1222 | 1242 | -0.090 | 0.035 | 0.173 | 0.171 | -0.022 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| MET |   | 6 | 301 | 1047 | 1020 | -0.219 | 0.086 | 0.183 | -0.025 | 0.170 |
|---|
| GARS |   | 2 | 743 | 1222 | 1215 | 0.087 | 0.149 | -0.237 | 0.171 | 0.045 |
|---|
| AKT1 |   | 16 | 104 | 236 | 227 | 0.045 | 0.126 | -0.099 | undef | -0.168 |
|---|
| UQCRQ |   | 2 | 743 | 1 | 66 | 0.171 | 0.035 | -0.031 | 0.141 | 0.010 |
|---|
| AKT2 |   | 28 | 47 | 412 | 394 | 0.110 | -0.118 | -0.003 | 0.088 | -0.092 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| DSP |   | 7 | 256 | 590 | 570 | 0.054 | 0.235 | -0.119 | -0.033 | 0.053 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| OPTN |   | 7 | 256 | 1 | 47 | 0.226 | -0.099 | 0.015 | undef | 0.035 |
|---|
| SH3RF1 |   | 6 | 301 | 1222 | 1194 | 0.144 | 0.238 | undef | undef | undef |
|---|
| NUPR1 |   | 3 | 557 | 1222 | 1210 | 0.057 | -0.031 | -0.241 | 0.145 | 0.013 |
|---|
| EIF4EBP1 |   | 3 | 557 | 1222 | 1210 | 0.086 | -0.201 | -0.147 | 0.183 | 0.216 |
|---|
| TRAF3IP1 |   | 9 | 196 | 83 | 99 | -0.113 | -0.063 | -0.088 | 0.276 | undef |
|---|
| PRDX5 |   | 1 | 1195 | 1222 | 1242 | 0.154 | -0.170 | undef | 0.145 | undef |
|---|
| FGF7 |   | 10 | 167 | 780 | 760 | -0.094 | 0.237 | 0.070 | 0.038 | 0.083 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| NRP1 |   | 8 | 222 | 1222 | 1186 | -0.043 | 0.025 | 0.201 | 0.029 | 0.071 |
|---|
| LTA4H |   | 1 | 1195 | 1222 | 1242 | 0.179 | -0.229 | 0.135 | -0.108 | -0.042 |
|---|
| ANGPT1 |   | 5 | 360 | 696 | 680 | 0.106 | -0.167 | 0.084 | -0.035 | 0.250 |
|---|
| PSTPIP1 |   | 1 | 1195 | 1222 | 1242 | 0.036 | -0.258 | -0.092 | -0.104 | -0.102 |
|---|
| TUBB |   | 5 | 360 | 614 | 612 | -0.118 | 0.170 | 0.200 | undef | -0.045 |
|---|
| PARD3 |   | 6 | 301 | 366 | 377 | 0.030 | -0.006 | -0.052 | 0.040 | -0.090 |
|---|
| FN1 |   | 6 | 301 | 590 | 582 | 0.190 | 0.133 | 0.091 | undef | 0.083 |
|---|
| ACTC1 |   | 2 | 743 | 1222 | 1215 | 0.185 | 0.066 | 0.115 | 0.138 | 0.076 |
|---|
| SDHC |   | 3 | 557 | 1141 | 1129 | 0.158 | -0.033 | -0.077 | 0.199 | 0.110 |
|---|
| SMAD7 |   | 5 | 360 | 1034 | 1004 | -0.035 | 0.131 | 0.169 | 0.061 | 0.147 |
|---|
| RACGAP1 |   | 4 | 440 | 1222 | 1205 | 0.283 | -0.164 | 0.073 | 0.055 | 0.188 |
|---|
| LIAS |   | 1 | 1195 | 1222 | 1242 | -0.024 | 0.045 | 0.085 | undef | -0.125 |
|---|
| CDH11 |   | 2 | 743 | 1222 | 1215 | -0.047 | 0.067 | -0.105 | undef | 0.074 |
|---|
| PNO1 |   | 3 | 557 | 1010 | 1001 | 0.090 | -0.052 | -0.092 | -0.048 | -0.138 |
|---|
| THBS1 |   | 5 | 360 | 1141 | 1109 | 0.078 | 0.044 | 0.185 | 0.200 | 0.267 |
|---|
| CAPN2 |   | 1 | 1195 | 1222 | 1242 | 0.031 | 0.156 | 0.042 | 0.186 | 0.082 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| PAK2 |   | 26 | 52 | 179 | 177 | 0.014 | 0.161 | 0.081 | -0.044 | 0.062 |
|---|
| C1orf9 |   | 1 | 1195 | 1222 | 1242 | 0.334 | 0.146 | -0.026 | 0.180 | 0.096 |
|---|
| FBXL2 |   | 2 | 743 | 1222 | 1215 | 0.151 | 0.089 | 0.089 | undef | -0.116 |
|---|
| MYB |   | 28 | 47 | 366 | 344 | -0.028 | 0.075 | 0.002 | 0.065 | -0.012 |
|---|
| PRSS23 |   | 1 | 1195 | 1222 | 1242 | 0.133 | 0.056 | 0.060 | 0.158 | 0.092 |
|---|
| RIPK3 |   | 19 | 85 | 179 | 186 | -0.014 | 0.040 | undef | 0.173 | undef |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| ITGA5 |   | 26 | 52 | 179 | 177 | 0.031 | 0.192 | 0.125 | 0.133 | 0.202 |
|---|
| COL7A1 |   | 3 | 557 | 1222 | 1210 | -0.052 | 0.240 | 0.233 | -0.019 | 0.022 |
|---|
| FANCA |   | 9 | 196 | 1 | 44 | 0.120 | -0.188 | 0.106 | -0.127 | -0.053 |
|---|
| MARK2 |   | 1 | 1195 | 1222 | 1242 | -0.085 | 0.130 | -0.129 | undef | -0.212 |
|---|
| IRS4 |   | 32 | 40 | 236 | 224 | 0.115 | -0.111 | -0.088 | 0.096 | 0.142 |
|---|
| CDK5R1 |   | 17 | 95 | 552 | 521 | -0.150 | 0.038 | -0.096 | 0.121 | 0.091 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| SOX13 |   | 4 | 440 | 1034 | 1015 | -0.113 | 0.095 | -0.002 | 0.271 | -0.184 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| IKBKE |   | 10 | 167 | 552 | 539 | 0.227 | -0.041 | -0.095 | -0.197 | 0.079 |
|---|
| COQ10A |   | 1 | 1195 | 1222 | 1242 | 0.039 | 0.091 | undef | undef | undef |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| YWHAZ |   | 3 | 557 | 658 | 654 | 0.043 | -0.274 | -0.018 | undef | 0.054 |
|---|
| DIMT1L |   | 1 | 1195 | 1222 | 1242 | 0.043 | -0.061 | 0.012 | -0.075 | 0.119 |
|---|
| ITGB4 |   | 35 | 34 | 1 | 20 | 0.021 | 0.313 | -0.134 | 0.080 | -0.114 |
|---|
| HEXA |   | 3 | 557 | 1141 | 1129 | 0.131 | -0.121 | 0.091 | -0.031 | -0.144 |
|---|
| FAM178A |   | 1 | 1195 | 1222 | 1242 | 0.221 | 0.112 | 0.226 | 0.212 | 0.200 |
|---|
GO Enrichment output for subnetwork 6885 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 5.142E-09 | 1.256E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.273E-07 | 1.555E-04 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 1.407E-07 | 1.146E-04 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 1.79E-07 | 1.093E-04 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 2.112E-07 | 1.032E-04 |
|---|
| regulation of behavior | GO:0050795 |  | 2.8E-07 | 1.14E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 3.303E-07 | 1.153E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 4.933E-07 | 1.506E-04 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 4.933E-07 | 1.339E-04 |
|---|
| cell killing | GO:0001906 |  | 1.342E-06 | 3.279E-04 |
|---|
| polyol catabolic process | GO:0046174 |  | 1.82E-06 | 4.042E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 6.71E-09 | 1.615E-05 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 1.515E-07 | 1.823E-04 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.659E-07 | 1.33E-04 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 1.951E-07 | 1.174E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 2.752E-07 | 1.324E-04 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 2.752E-07 | 1.103E-04 |
|---|
| regulation of behavior | GO:0050795 |  | 3.118E-07 | 1.072E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 6.424E-07 | 1.932E-04 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 6.424E-07 | 1.717E-04 |
|---|
| cell killing | GO:0001906 |  | 1.746E-06 | 4.202E-04 |
|---|
| regulation of alternative nuclear mRNA splicing. via spliceosome | GO:0000381 |  | 2.223E-06 | 4.863E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 2.384E-08 | 5.483E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 3.272E-07 | 3.763E-04 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 5.499E-07 | 4.216E-04 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 5.852E-07 | 3.365E-04 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 7.168E-07 | 3.297E-04 |
|---|
| regulation of behavior | GO:0050795 |  | 9.214E-07 | 3.532E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 1.513E-06 | 4.97E-04 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 1.513E-06 | 4.349E-04 |
|---|
| cell killing | GO:0001906 |  | 4.499E-06 | 1.15E-03 |
|---|
| polyol catabolic process | GO:0046174 |  | 5.754E-06 | 1.323E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 5.754E-06 | 1.203E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 3.362E-06 | 6.196E-03 |
|---|
| angiogenesis | GO:0001525 |  | 5.222E-06 | 4.812E-03 |
|---|
| positive regulation of endothelial cell migration | GO:0010595 |  | 1.855E-05 | 0.01139292 |
|---|
| positive regulation of leukocyte migration | GO:0002687 |  | 1.855E-05 | 8.545E-03 |
|---|
| blood vessel morphogenesis | GO:0048514 |  | 2.373E-05 | 8.748E-03 |
|---|
| regulation of leukocyte migration | GO:0002685 |  | 2.768E-05 | 8.502E-03 |
|---|
| regulation of blood vessel endothelial cell migration | GO:0043535 |  | 2.768E-05 | 7.287E-03 |
|---|
| heart development | GO:0007507 |  | 3.108E-05 | 7.159E-03 |
|---|
| response to oxygen levels | GO:0070482 |  | 3.618E-05 | 7.41E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 4.101E-05 | 7.558E-03 |
|---|
| blood vessel development | GO:0001568 |  | 4.826E-05 | 8.085E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 2.384E-08 | 5.483E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 3.272E-07 | 3.763E-04 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 5.499E-07 | 4.216E-04 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 5.852E-07 | 3.365E-04 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 7.168E-07 | 3.297E-04 |
|---|
| regulation of behavior | GO:0050795 |  | 9.214E-07 | 3.532E-04 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 1.513E-06 | 4.97E-04 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 1.513E-06 | 4.349E-04 |
|---|
| cell killing | GO:0001906 |  | 4.499E-06 | 1.15E-03 |
|---|
| polyol catabolic process | GO:0046174 |  | 5.754E-06 | 1.323E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 5.754E-06 | 1.203E-03 |
|---|



