Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6884
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7490 | 3.834e-03 | 6.400e-04 | 7.826e-01 | 1.920e-06 |
|---|
| Loi | 0.2309 | 7.933e-02 | 1.111e-02 | 4.259e-01 | 3.755e-04 |
|---|
| Schmidt | 0.6760 | 0.000e+00 | 0.000e+00 | 4.216e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7911 | 0.000e+00 | 0.000e+00 | 3.425e-03 | 0.000e+00 |
|---|
| Wang | 0.2636 | 2.414e-03 | 4.135e-02 | 3.846e-01 | 3.839e-05 |
|---|
Expression data for subnetwork 6884 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
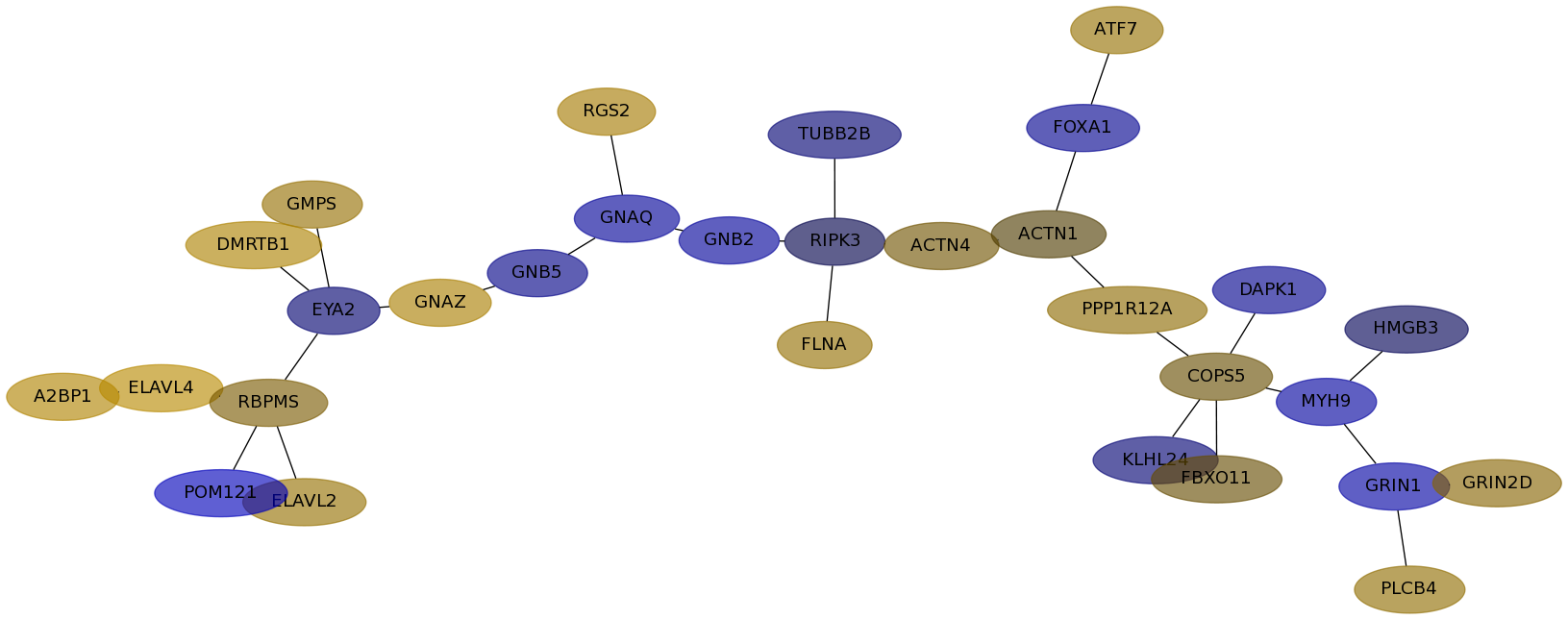
Score for each gene in subnetwork 6884 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| KLHL24 |   | 4 | 440 | 682 | 678 | -0.036 | -0.011 | 0.062 | 0.044 | -0.125 |
|---|
| PPP1R12A |   | 2 | 743 | 318 | 346 | 0.069 | 0.124 | 0.114 | 0.100 | 0.018 |
|---|
| RIPK3 |   | 19 | 85 | 179 | 186 | -0.014 | 0.040 | undef | 0.173 | undef |
|---|
| A2BP1 |   | 1 | 1195 | 1131 | 1158 | 0.156 | 0.034 | 0.000 | -0.127 | undef |
|---|
| ATF7 |   | 28 | 47 | 179 | 175 | 0.085 | 0.115 | -0.088 | 0.172 | -0.092 |
|---|
| GRIN1 |   | 3 | 557 | 928 | 925 | -0.114 | -0.067 | -0.111 | undef | 0.115 |
|---|
| MYH9 |   | 4 | 440 | 1131 | 1108 | -0.103 | 0.227 | 0.007 | -0.150 | 0.165 |
|---|
| ACTN1 |   | 18 | 86 | 318 | 313 | 0.016 | 0.347 | 0.068 | 0.100 | 0.314 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| RGS2 |   | 3 | 557 | 1131 | 1126 | 0.118 | 0.061 | 0.084 | 0.050 | 0.178 |
|---|
| GNB2 |   | 11 | 148 | 525 | 505 | -0.091 | 0.082 | -0.062 | 0.208 | -0.062 |
|---|
| POM121 |   | 7 | 256 | 780 | 763 | -0.197 | 0.101 | 0.079 | undef | 0.017 |
|---|
| ELAVL4 |   | 2 | 743 | 1131 | 1136 | 0.187 | 0.047 | -0.088 | undef | -0.085 |
|---|
| FBXO11 |   | 5 | 360 | 457 | 455 | 0.027 | 0.186 | -0.069 | 0.238 | 0.149 |
|---|
| ELAVL2 |   | 1 | 1195 | 1131 | 1158 | 0.085 | 0.115 | 0.158 | undef | -0.076 |
|---|
| GNB5 |   | 9 | 196 | 1131 | 1100 | -0.059 | 0.026 | 0.064 | 0.284 | 0.103 |
|---|
| GNAZ |   | 15 | 111 | 478 | 457 | 0.134 | -0.116 | 0.308 | 0.267 | 0.011 |
|---|
| COPS5 |   | 2 | 743 | 1131 | 1136 | 0.028 | -0.117 | -0.030 | 0.155 | 0.108 |
|---|
| DAPK1 |   | 1 | 1195 | 1131 | 1158 | -0.067 | 0.041 | 0.033 | -0.170 | 0.009 |
|---|
| TUBB2B |   | 13 | 124 | 141 | 141 | -0.036 | 0.304 | 0.045 | 0.187 | 0.139 |
|---|
| PLCB4 |   | 4 | 440 | 928 | 912 | 0.074 | 0.179 | -0.113 | 0.248 | -0.020 |
|---|
| GMPS |   | 7 | 256 | 318 | 328 | 0.083 | -0.022 | 0.044 | -0.019 | 0.151 |
|---|
| ACTN4 |   | 14 | 117 | 318 | 315 | 0.035 | 0.180 | undef | -0.048 | undef |
|---|
| GNAQ |   | 2 | 743 | 525 | 547 | -0.090 | 0.226 | 0.105 | undef | -0.062 |
|---|
| GRIN2D |   | 13 | 124 | 552 | 522 | 0.058 | 0.155 | 0.000 | undef | undef |
|---|
| DMRTB1 |   | 1 | 1195 | 1131 | 1158 | 0.145 | 0.078 | undef | undef | undef |
|---|
| RBPMS |   | 3 | 557 | 1010 | 1001 | 0.043 | 0.108 | 0.066 | 0.145 | -0.077 |
|---|
| EYA2 |   | 13 | 124 | 318 | 316 | -0.034 | 0.113 | -0.142 | undef | -0.019 |
|---|
| FOXA1 |   | 27 | 50 | 179 | 176 | -0.072 | 0.071 | -0.015 | 0.214 | 0.005 |
|---|
| HMGB3 |   | 1 | 1195 | 1131 | 1158 | -0.018 | -0.070 | -0.059 | 0.148 | 0.063 |
|---|
GO Enrichment output for subnetwork 6884 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glutamate signaling pathway | GO:0007215 |  | 2.368E-07 | 5.784E-04 |
|---|
| dopamine receptor signaling pathway | GO:0007212 |  | 3.067E-06 | 3.746E-03 |
|---|
| regulation of excitatory postsynaptic membrane potential | GO:0060079 |  | 6.309E-06 | 5.137E-03 |
|---|
| actin cytoskeleton reorganization | GO:0031532 |  | 6.309E-06 | 3.853E-03 |
|---|
| regulation of postsynaptic membrane potential | GO:0060078 |  | 1.334E-05 | 6.518E-03 |
|---|
| visual learning | GO:0008542 |  | 1.334E-05 | 5.432E-03 |
|---|
| actin filament bundle formation | GO:0051017 |  | 1.567E-05 | 5.468E-03 |
|---|
| visual behavior | GO:0007632 |  | 2.421E-05 | 7.393E-03 |
|---|
| response to ethanol | GO:0045471 |  | 2.421E-05 | 6.572E-03 |
|---|
| membrane depolarization | GO:0051899 |  | 2.762E-05 | 6.748E-03 |
|---|
| actin filament organization | GO:0007015 |  | 3.033E-05 | 6.737E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glutamate signaling pathway | GO:0007215 |  | 2.789E-07 | 6.711E-04 |
|---|
| dopamine receptor signaling pathway | GO:0007212 |  | 3.477E-06 | 4.183E-03 |
|---|
| actin cytoskeleton reorganization | GO:0031532 |  | 5.731E-06 | 4.597E-03 |
|---|
| regulation of excitatory postsynaptic membrane potential | GO:0060079 |  | 7.151E-06 | 4.301E-03 |
|---|
| regulation of postsynaptic membrane potential | GO:0060078 |  | 1.512E-05 | 7.275E-03 |
|---|
| actin filament bundle formation | GO:0051017 |  | 1.512E-05 | 6.062E-03 |
|---|
| visual learning | GO:0008542 |  | 1.512E-05 | 5.196E-03 |
|---|
| actin filament organization | GO:0007015 |  | 2.524E-05 | 7.59E-03 |
|---|
| visual behavior | GO:0007632 |  | 2.743E-05 | 7.332E-03 |
|---|
| response to ethanol | GO:0045471 |  | 2.743E-05 | 6.599E-03 |
|---|
| membrane depolarization | GO:0051899 |  | 3.129E-05 | 6.844E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glutamate signaling pathway | GO:0007215 |  | 3.839E-07 | 8.829E-04 |
|---|
| dopamine receptor signaling pathway | GO:0007212 |  | 3.361E-06 | 3.865E-03 |
|---|
| actin cytoskeleton reorganization | GO:0031532 |  | 3.361E-06 | 2.577E-03 |
|---|
| regulation of excitatory postsynaptic membrane potential | GO:0060079 |  | 9.195E-06 | 5.287E-03 |
|---|
| visual learning | GO:0008542 |  | 1.639E-05 | 7.541E-03 |
|---|
| regulation of postsynaptic membrane potential | GO:0060078 |  | 1.943E-05 | 7.448E-03 |
|---|
| response to ethanol | GO:0045471 |  | 2.281E-05 | 7.495E-03 |
|---|
| visual behavior | GO:0007632 |  | 2.656E-05 | 7.636E-03 |
|---|
| membrane depolarization | GO:0051899 |  | 3.069E-05 | 7.844E-03 |
|---|
| forebrain neuron development | GO:0021884 |  | 7.655E-05 | 0.0176064 |
|---|
| early endosome to late endosome transport | GO:0045022 |  | 7.655E-05 | 0.01600582 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| prostate gland development | GO:0030850 |  | 7.191E-05 | 0.1325224 |
|---|
| dopamine receptor signaling pathway | GO:0007212 |  | 7.191E-05 | 0.0662612 |
|---|
| receptor clustering | GO:0043113 |  | 7.191E-05 | 0.04417413 |
|---|
| actin cytoskeleton reorganization | GO:0031532 |  | 7.191E-05 | 0.0331306 |
|---|
| positive regulation of transcription factor import into nucleus | GO:0042993 |  | 1.077E-04 | 0.03969123 |
|---|
| purine base biosynthetic process | GO:0009113 |  | 1.077E-04 | 0.03307602 |
|---|
| negative regulation of blood coagulation | GO:0030195 |  | 2.003E-04 | 0.05274745 |
|---|
| cytoplasmic sequestering of protein | GO:0051220 |  | 2.003E-04 | 0.04615402 |
|---|
| glutamine metabolic process | GO:0006541 |  | 2.572E-04 | 0.05266059 |
|---|
| nucleobase biosynthetic process | GO:0046112 |  | 2.572E-04 | 0.04739453 |
|---|
| positive regulation of protein import into nucleus | GO:0042307 |  | 2.572E-04 | 0.04308594 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glutamate signaling pathway | GO:0007215 |  | 3.839E-07 | 8.829E-04 |
|---|
| dopamine receptor signaling pathway | GO:0007212 |  | 3.361E-06 | 3.865E-03 |
|---|
| actin cytoskeleton reorganization | GO:0031532 |  | 3.361E-06 | 2.577E-03 |
|---|
| regulation of excitatory postsynaptic membrane potential | GO:0060079 |  | 9.195E-06 | 5.287E-03 |
|---|
| visual learning | GO:0008542 |  | 1.639E-05 | 7.541E-03 |
|---|
| regulation of postsynaptic membrane potential | GO:0060078 |  | 1.943E-05 | 7.448E-03 |
|---|
| response to ethanol | GO:0045471 |  | 2.281E-05 | 7.495E-03 |
|---|
| visual behavior | GO:0007632 |  | 2.656E-05 | 7.636E-03 |
|---|
| membrane depolarization | GO:0051899 |  | 3.069E-05 | 7.844E-03 |
|---|
| forebrain neuron development | GO:0021884 |  | 7.655E-05 | 0.0176064 |
|---|
| early endosome to late endosome transport | GO:0045022 |  | 7.655E-05 | 0.01600582 |
|---|



