Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6849
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7493 | 3.815e-03 | 6.350e-04 | 7.819e-01 | 1.894e-06 |
|---|
| Loi | 0.2308 | 7.955e-02 | 1.117e-02 | 4.265e-01 | 3.789e-04 |
|---|
| Schmidt | 0.6758 | 0.000e+00 | 0.000e+00 | 4.238e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7911 | 0.000e+00 | 0.000e+00 | 3.428e-03 | 0.000e+00 |
|---|
| Wang | 0.2635 | 2.424e-03 | 4.144e-02 | 3.851e-01 | 3.868e-05 |
|---|
Expression data for subnetwork 6849 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
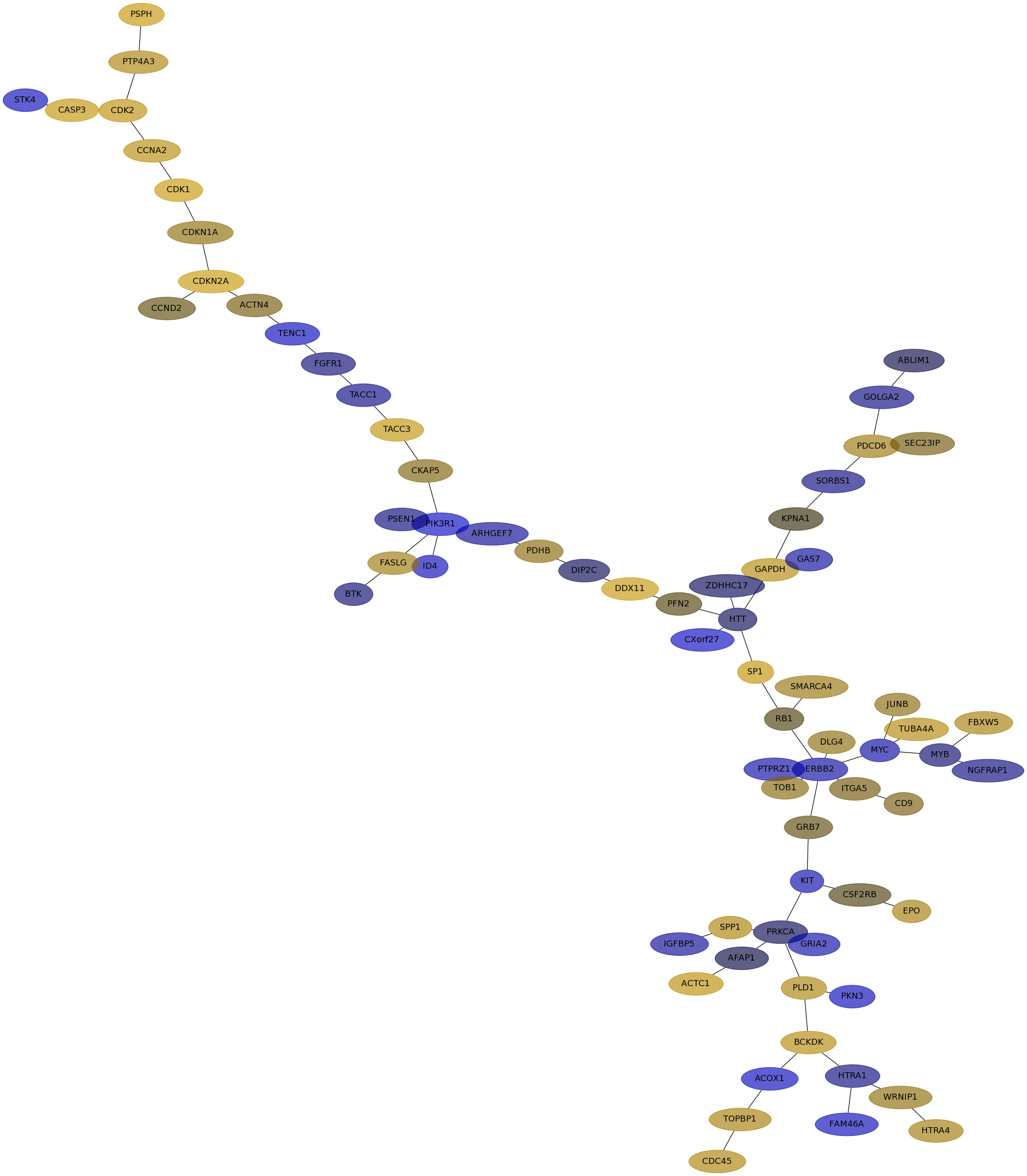
Score for each gene in subnetwork 6849 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| CASP3 |   | 11 | 148 | 141 | 143 | 0.234 | -0.032 | -0.142 | -0.097 | -0.084 |
|---|
| ZDHHC17 |   | 1 | 1195 | 1492 | 1518 | -0.018 | 0.074 | 0.121 | undef | -0.052 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| HTRA1 |   | 7 | 256 | 83 | 102 | -0.046 | 0.184 | 0.051 | 0.132 | 0.224 |
|---|
| SMARCA4 |   | 4 | 440 | 1364 | 1330 | 0.080 | 0.202 | -0.071 | -0.050 | -0.116 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| PTP4A3 |   | 9 | 196 | 412 | 411 | 0.137 | -0.196 | 0.085 | undef | -0.004 |
|---|
| ID4 |   | 35 | 34 | 236 | 223 | -0.179 | 0.137 | 0.241 | undef | 0.036 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| DIP2C |   | 1 | 1195 | 1492 | 1518 | -0.017 | 0.046 | 0.039 | 0.200 | 0.081 |
|---|
| CCNA2 |   | 21 | 73 | 179 | 183 | 0.176 | -0.053 | 0.188 | -0.078 | 0.246 |
|---|
| FAM46A |   | 2 | 743 | 1351 | 1344 | -0.177 | 0.090 | undef | 0.125 | undef |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| TACC3 |   | 4 | 440 | 1077 | 1065 | 0.232 | -0.144 | 0.014 | 0.046 | 0.004 |
|---|
| TUBA4A |   | 7 | 256 | 1 | 47 | 0.157 | 0.136 | -0.010 | undef | 0.156 |
|---|
| GAPDH |   | 11 | 148 | 141 | 143 | 0.159 | -0.040 | -0.053 | 0.088 | 0.029 |
|---|
| BTK |   | 2 | 743 | 1492 | 1474 | -0.034 | -0.210 | -0.081 | -0.218 | -0.046 |
|---|
| DLG4 |   | 5 | 360 | 806 | 784 | 0.059 | -0.113 | 0.151 | -0.131 | 0.123 |
|---|
| PDHB |   | 13 | 124 | 525 | 503 | 0.056 | 0.017 | 0.197 | -0.205 | 0.193 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| FGFR1 |   | 26 | 52 | 638 | 615 | -0.036 | 0.200 | 0.087 | undef | -0.015 |
|---|
| CDC45 |   | 5 | 360 | 318 | 334 | 0.134 | -0.171 | 0.185 | -0.103 | -0.085 |
|---|
| NGFRAP1 |   | 3 | 557 | 991 | 982 | -0.044 | 0.005 | 0.252 | -0.001 | 0.109 |
|---|
| HTRA4 |   | 8 | 222 | 83 | 101 | 0.102 | undef | undef | undef | undef |
|---|
| ACTN4 |   | 14 | 117 | 318 | 315 | 0.035 | 0.180 | undef | -0.048 | undef |
|---|
| PSPH |   | 20 | 81 | 366 | 356 | 0.240 | -0.019 | -0.102 | 0.358 | 0.025 |
|---|
| BCKDK |   | 3 | 557 | 83 | 117 | 0.166 | -0.004 | -0.012 | -0.031 | -0.001 |
|---|
| ACTC1 |   | 2 | 743 | 1222 | 1215 | 0.185 | 0.066 | 0.115 | 0.138 | 0.076 |
|---|
| GAS7 |   | 3 | 557 | 83 | 117 | -0.093 | -0.082 | 0.172 | 0.120 | 0.064 |
|---|
| DDX11 |   | 3 | 557 | 179 | 213 | 0.254 | -0.126 | -0.191 | -0.033 | -0.135 |
|---|
| ARHGEF7 |   | 25 | 59 | 83 | 92 | -0.074 | -0.061 | 0.208 | 0.028 | 0.015 |
|---|
| GRIA2 |   | 2 | 743 | 1492 | 1474 | -0.115 | -0.043 | 0.000 | -0.066 | 0.121 |
|---|
| PSEN1 |   | 10 | 167 | 366 | 362 | -0.041 | 0.110 | undef | 0.083 | undef |
|---|
| PFN2 |   | 11 | 148 | 141 | 143 | 0.014 | -0.001 | 0.286 | -0.002 | -0.056 |
|---|
| CKAP5 |   | 1 | 1195 | 1492 | 1518 | 0.044 | -0.069 | -0.035 | undef | 0.023 |
|---|
| WRNIP1 |   | 3 | 557 | 83 | 117 | 0.064 | -0.025 | 0.162 | undef | -0.115 |
|---|
| CSF2RB |   | 3 | 557 | 1492 | 1464 | 0.013 | -0.196 | -0.109 | 0.012 | -0.141 |
|---|
| ABLIM1 |   | 1 | 1195 | 1492 | 1518 | -0.011 | 0.066 | 0.169 | 0.181 | 0.061 |
|---|
| RB1 |   | 31 | 43 | 1 | 22 | 0.013 | -0.051 | 0.057 | 0.100 | -0.082 |
|---|
| SPP1 |   | 26 | 52 | 83 | 91 | 0.130 | 0.119 | 0.184 | 0.204 | 0.153 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| JUNB |   | 20 | 81 | 179 | 185 | 0.057 | 0.086 | 0.065 | 0.001 | -0.015 |
|---|
| CD9 |   | 13 | 124 | 525 | 503 | 0.036 | -0.115 | -0.109 | 0.281 | -0.147 |
|---|
| PTPRZ1 |   | 9 | 196 | 570 | 554 | -0.114 | 0.183 | 0.233 | -0.316 | -0.095 |
|---|
| CCND2 |   | 5 | 360 | 1492 | 1453 | 0.022 | -0.011 | -0.072 | -0.153 | -0.029 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| MYB |   | 28 | 47 | 366 | 344 | -0.028 | 0.075 | 0.002 | 0.065 | -0.012 |
|---|
| TOPBP1 |   | 1 | 1195 | 1492 | 1518 | 0.118 | -0.044 | 0.116 | undef | 0.084 |
|---|
| CXorf27 |   | 1 | 1195 | 1492 | 1518 | -0.214 | -0.088 | 0.000 | -0.155 | 0.153 |
|---|
| TACC1 |   | 5 | 360 | 1077 | 1057 | -0.051 | 0.020 | -0.022 | 0.187 | 0.126 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| ITGA5 |   | 26 | 52 | 179 | 177 | 0.031 | 0.192 | 0.125 | 0.133 | 0.202 |
|---|
| TENC1 |   | 2 | 743 | 1492 | 1474 | -0.194 | 0.149 | 0.093 | undef | -0.159 |
|---|
| SEC23IP |   | 1 | 1195 | 1492 | 1518 | 0.033 | -0.006 | -0.033 | 0.106 | -0.055 |
|---|
| KPNA1 |   | 9 | 196 | 141 | 147 | 0.007 | 0.152 | -0.083 | 0.073 | 0.086 |
|---|
| PDCD6 |   | 10 | 167 | 1 | 40 | 0.091 | -0.060 | undef | undef | undef |
|---|
| GOLGA2 |   | 2 | 743 | 1492 | 1474 | -0.048 | 0.213 | 0.050 | undef | 0.052 |
|---|
| PKN3 |   | 18 | 86 | 179 | 187 | -0.181 | 0.009 | undef | 0.239 | undef |
|---|
| HTT |   | 14 | 117 | 141 | 140 | -0.016 | -0.015 | 0.075 | 0.092 | -0.093 |
|---|
| ACOX1 |   | 9 | 196 | 1 | 44 | -0.209 | 0.122 | -0.104 | 0.252 | -0.094 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| FASLG |   | 1 | 1195 | 1492 | 1518 | 0.092 | -0.227 | -0.138 | -0.178 | -0.174 |
|---|
| EPO |   | 4 | 440 | 1492 | 1461 | 0.109 | 0.086 | -0.083 | -0.048 | 0.155 |
|---|
| PLD1 |   | 17 | 95 | 1 | 29 | 0.132 | 0.033 | -0.158 | -0.062 | 0.123 |
|---|
| CDK2 |   | 31 | 43 | 179 | 174 | 0.196 | -0.096 | 0.109 | 0.001 | 0.362 |
|---|
| TOB1 |   | 6 | 301 | 525 | 517 | 0.055 | 0.097 | -0.069 | 0.107 | -0.081 |
|---|
| AFAP1 |   | 2 | 743 | 1492 | 1474 | -0.010 | -0.051 | -0.050 | undef | 0.052 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
| FBXW5 |   | 1 | 1195 | 1492 | 1518 | 0.113 | 0.077 | undef | undef | undef |
|---|
| STK4 |   | 1 | 1195 | 1492 | 1518 | -0.177 | -0.270 | 0.244 | 0.058 | 0.152 |
|---|
GO Enrichment output for subnetwork 6849 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.374E-12 | 1.557E-08 |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 6.711E-12 | 8.198E-09 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 5.323E-11 | 4.335E-08 |
|---|
| S phase | GO:0051320 |  | 5.404E-10 | 3.301E-07 |
|---|
| mitotic sister chromatid segregation | GO:0000070 |  | 3.91E-08 | 1.91E-05 |
|---|
| sister chromatid segregation | GO:0000819 |  | 4.646E-08 | 1.892E-05 |
|---|
| chromosome segregation | GO:0007059 |  | 7.953E-08 | 2.775E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 9.777E-08 | 2.986E-05 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.081E-07 | 2.935E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.57E-07 | 3.836E-05 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 1.594E-07 | 3.54E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.095E-12 | 9.853E-09 |
|---|
| interphase | GO:0051325 |  | 5.795E-12 | 6.972E-09 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 5.067E-11 | 4.064E-08 |
|---|
| S phase | GO:0051320 |  | 5.241E-10 | 3.152E-07 |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 8.126E-09 | 3.91E-06 |
|---|
| mitotic sister chromatid segregation | GO:0000070 |  | 3.762E-08 | 1.508E-05 |
|---|
| sister chromatid segregation | GO:0000819 |  | 4.47E-08 | 1.536E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 7.586E-08 | 2.282E-05 |
|---|
| chromosome segregation | GO:0007059 |  | 7.586E-08 | 2.028E-05 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 8.419E-08 | 2.026E-05 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 1.256E-07 | 2.748E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.016E-10 | 1.154E-06 |
|---|
| interphase | GO:0051325 |  | 6.319E-10 | 7.266E-07 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 5.607E-09 | 4.299E-06 |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 2.347E-08 | 1.35E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.501E-07 | 6.903E-05 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 1.651E-07 | 6.329E-05 |
|---|
| S phase | GO:0051320 |  | 1.651E-07 | 5.425E-05 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.697E-07 | 4.878E-05 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 2.419E-07 | 6.181E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.553E-07 | 8.171E-05 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 4.899E-07 | 1.024E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 5.572E-07 | 1.027E-03 |
|---|
| cell junction assembly | GO:0034329 |  | 5.938E-06 | 5.472E-03 |
|---|
| negative regulation of T cell proliferation | GO:0042130 |  | 7.557E-06 | 4.643E-03 |
|---|
| negative regulation of BMP signaling pathway | GO:0030514 |  | 7.557E-06 | 3.482E-03 |
|---|
| negative regulation of lymphocyte proliferation | GO:0050672 |  | 1.316E-05 | 4.849E-03 |
|---|
| negative regulation of T cell activation | GO:0050868 |  | 1.316E-05 | 4.041E-03 |
|---|
| cell junction organization | GO:0034330 |  | 1.776E-05 | 4.675E-03 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 3.018E-05 | 6.952E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.327E-05 | 6.813E-03 |
|---|
| induction of apoptosis via death domain receptors | GO:0008625 |  | 4.44E-05 | 8.183E-03 |
|---|
| cell-substrate junction assembly | GO:0007044 |  | 4.44E-05 | 7.439E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.016E-10 | 1.154E-06 |
|---|
| interphase | GO:0051325 |  | 6.319E-10 | 7.266E-07 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 5.607E-09 | 4.299E-06 |
|---|
| induction of apoptosis by extracellular signals | GO:0008624 |  | 2.347E-08 | 1.35E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.501E-07 | 6.903E-05 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 1.651E-07 | 6.329E-05 |
|---|
| S phase | GO:0051320 |  | 1.651E-07 | 5.425E-05 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.697E-07 | 4.878E-05 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 2.419E-07 | 6.181E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.553E-07 | 8.171E-05 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 4.899E-07 | 1.024E-04 |
|---|



