Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6763
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7501 | 3.756e-03 | 6.230e-04 | 7.799e-01 | 1.825e-06 |
|---|
| Loi | 0.2309 | 7.933e-02 | 1.111e-02 | 4.259e-01 | 3.755e-04 |
|---|
| Schmidt | 0.6749 | 0.000e+00 | 0.000e+00 | 4.331e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7911 | 0.000e+00 | 0.000e+00 | 3.428e-03 | 0.000e+00 |
|---|
| Wang | 0.2633 | 2.456e-03 | 4.175e-02 | 3.866e-01 | 3.964e-05 |
|---|
Expression data for subnetwork 6763 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
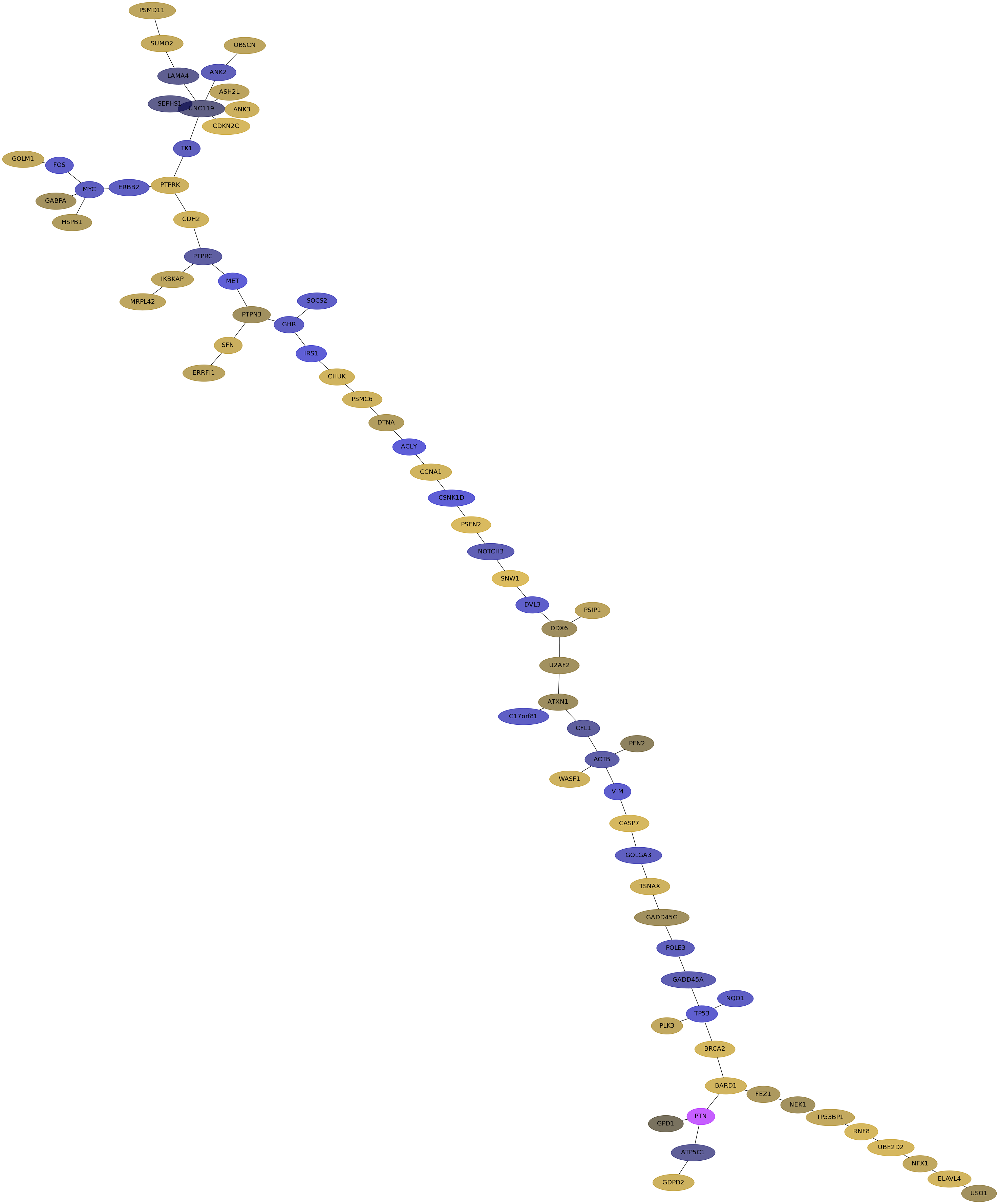
Score for each gene in subnetwork 6763 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| NQO1 |   | 21 | 73 | 366 | 354 | -0.115 | 0.069 | -0.189 | 0.321 | 0.118 |
|---|
| WASF1 |   | 7 | 256 | 1 | 47 | 0.162 | -0.070 | 0.348 | -0.037 | 0.130 |
|---|
| SUMO2 |   | 22 | 69 | 366 | 345 | 0.117 | 0.062 | 0.294 | 0.141 | -0.096 |
|---|
| ANK3 |   | 6 | 301 | 83 | 105 | 0.153 | 0.034 | 0.001 | 0.094 | -0.038 |
|---|
| SOCS2 |   | 3 | 557 | 1688 | 1660 | -0.130 | -0.028 | 0.051 | -0.171 | 0.027 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CHUK |   | 6 | 301 | 318 | 330 | 0.173 | 0.080 | 0.152 | undef | 0.215 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| CDKN2C |   | 16 | 104 | 236 | 227 | 0.216 | -0.106 | 0.202 | -0.024 | -0.081 |
|---|
| GOLGA3 |   | 1 | 1195 | 1688 | 1703 | -0.094 | 0.240 | -0.084 | 0.145 | 0.137 |
|---|
| PSEN2 |   | 14 | 117 | 614 | 591 | 0.239 | -0.068 | -0.157 | 0.295 | 0.045 |
|---|
| GDPD2 |   | 1 | 1195 | 1688 | 1703 | 0.159 | -0.021 | 0.049 | -0.013 | -0.010 |
|---|
| ASH2L |   | 1 | 1195 | 1688 | 1703 | 0.084 | -0.064 | -0.124 | -0.016 | 0.074 |
|---|
| POLE3 |   | 17 | 95 | 141 | 138 | -0.083 | -0.033 | 0.231 | 0.096 | 0.011 |
|---|
| NOTCH3 |   | 5 | 360 | 806 | 784 | -0.063 | 0.115 | 0.136 | undef | -0.082 |
|---|
| MET |   | 6 | 301 | 1047 | 1020 | -0.219 | 0.086 | 0.183 | -0.025 | 0.170 |
|---|
| CFL1 |   | 2 | 743 | 1688 | 1673 | -0.027 | 0.087 | 0.010 | 0.303 | 0.008 |
|---|
| PTPRK |   | 14 | 117 | 83 | 96 | 0.156 | 0.141 | -0.094 | 0.048 | -0.024 |
|---|
| SEPHS1 |   | 3 | 557 | 957 | 949 | -0.014 | -0.311 | 0.129 | -0.023 | 0.033 |
|---|
| U2AF2 |   | 2 | 743 | 1260 | 1256 | 0.031 | 0.113 | 0.171 | -0.075 | 0.082 |
|---|
| LAMA4 |   | 13 | 124 | 696 | 670 | -0.016 | 0.138 | 0.209 | 0.172 | 0.343 |
|---|
| PLK3 |   | 2 | 743 | 1688 | 1673 | 0.101 | 0.053 | 0.038 | -0.091 | -0.002 |
|---|
| USO1 |   | 2 | 743 | 1688 | 1673 | 0.031 | -0.003 | -0.027 | 0.096 | -0.077 |
|---|
| GABPA |   | 23 | 66 | 696 | 665 | 0.033 | -0.183 | 0.066 | 0.141 | -0.161 |
|---|
| ANK2 |   | 3 | 557 | 1604 | 1577 | -0.073 | -0.023 | 0.095 | -0.019 | 0.095 |
|---|
| FEZ1 |   | 8 | 222 | 141 | 149 | 0.047 | -0.020 | 0.154 | 0.165 | -0.128 |
|---|
| ATXN1 |   | 17 | 95 | 1 | 29 | 0.026 | 0.192 | 0.175 | 0.125 | 0.163 |
|---|
| DVL3 |   | 1 | 1195 | 1688 | 1703 | -0.137 | 0.074 | 0.137 | 0.060 | -0.049 |
|---|
| VIM |   | 1 | 1195 | 1688 | 1703 | -0.159 | 0.075 | 0.207 | 0.008 | 0.015 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| PSMC6 |   | 5 | 360 | 318 | 334 | 0.163 | 0.061 | -0.010 | 0.069 | 0.044 |
|---|
| TP53BP1 |   | 1 | 1195 | 1688 | 1703 | 0.106 | 0.118 | 0.082 | -0.073 | 0.196 |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| ACTB |   | 3 | 557 | 1274 | 1258 | -0.039 | 0.283 | -0.061 | 0.164 | 0.003 |
|---|
| HSPB1 |   | 22 | 69 | 1 | 25 | 0.053 | 0.169 | 0.017 | 0.179 | 0.161 |
|---|
| PTN |   | 24 | 62 | 236 | 225 | undef | 0.022 | 0.000 | 0.054 | -0.092 |
|---|
| CSNK1D |   | 6 | 301 | 614 | 602 | -0.222 | -0.044 | 0.170 | -0.088 | 0.013 |
|---|
| MRPL42 |   | 1 | 1195 | 1688 | 1703 | 0.089 | -0.173 | 0.013 | -0.077 | -0.065 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| NFX1 |   | 1 | 1195 | 1688 | 1703 | 0.101 | -0.053 | -0.138 | -0.111 | 0.048 |
|---|
| PTPN3 |   | 11 | 148 | 658 | 635 | 0.029 | 0.060 | -0.067 | 0.225 | -0.034 |
|---|
| IKBKAP |   | 10 | 167 | 412 | 408 | 0.089 | 0.184 | 0.142 | 0.105 | -0.017 |
|---|
| PFN2 |   | 11 | 148 | 141 | 143 | 0.014 | -0.001 | 0.286 | -0.002 | -0.056 |
|---|
| PSIP1 |   | 1 | 1195 | 1688 | 1703 | 0.081 | -0.276 | 0.220 | undef | 0.126 |
|---|
| ELAVL4 |   | 2 | 743 | 1131 | 1136 | 0.187 | 0.047 | -0.088 | undef | -0.085 |
|---|
| GPD1 |   | 7 | 256 | 658 | 644 | 0.006 | -0.019 | 0.112 | -0.034 | 0.033 |
|---|
| GOLM1 |   | 8 | 222 | 1534 | 1489 | 0.109 | 0.161 | -0.217 | 0.170 | -0.060 |
|---|
| UNC119 |   | 10 | 167 | 83 | 98 | -0.009 | -0.324 | -0.099 | 0.094 | 0.071 |
|---|
| SNW1 |   | 1 | 1195 | 1688 | 1703 | 0.271 | 0.051 | 0.169 | -0.049 | 0.117 |
|---|
| TSNAX |   | 1 | 1195 | 1688 | 1703 | 0.162 | 0.075 | 0.018 | 0.209 | 0.154 |
|---|
| C17orf81 |   | 2 | 743 | 1674 | 1659 | -0.115 | -0.013 | -0.201 | -0.047 | 0.356 |
|---|
| DDX6 |   | 4 | 440 | 1688 | 1656 | 0.028 | 0.162 | 0.157 | -0.035 | 0.084 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| PSMD11 |   | 6 | 301 | 1437 | 1408 | 0.088 | -0.090 | -0.137 | 0.060 | -0.078 |
|---|
| ERRFI1 |   | 10 | 167 | 412 | 408 | 0.076 | 0.215 | undef | -0.043 | undef |
|---|
| TK1 |   | 4 | 440 | 727 | 723 | -0.089 | -0.156 | 0.029 | 0.119 | -0.052 |
|---|
| DTNA |   | 1 | 1195 | 1688 | 1703 | 0.058 | 0.073 | 0.276 | 0.006 | -0.016 |
|---|
| OBSCN |   | 1 | 1195 | 1688 | 1703 | 0.086 | 0.090 | -0.128 | undef | 0.095 |
|---|
| RNF8 |   | 2 | 743 | 1688 | 1673 | 0.216 | 0.004 | 0.114 | -0.088 | 0.221 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| PTPRC |   | 9 | 196 | 83 | 99 | -0.031 | -0.204 | -0.050 | -0.179 | -0.068 |
|---|
| IRS1 |   | 9 | 196 | 842 | 805 | -0.219 | 0.065 | 0.165 | 0.102 | 0.064 |
|---|
| CASP7 |   | 6 | 301 | 614 | 602 | 0.216 | 0.095 | 0.255 | undef | 0.035 |
|---|
| NEK1 |   | 15 | 111 | 141 | 139 | 0.032 | 0.184 | 0.049 | undef | 0.135 |
|---|
| GADD45A |   | 11 | 148 | 1351 | 1308 | -0.057 | 0.076 | -0.099 | 0.109 | 0.093 |
|---|
| ATP5C1 |   | 1 | 1195 | 1688 | 1703 | -0.022 | -0.047 | -0.030 | -0.121 | 0.146 |
|---|
| ACLY |   | 6 | 301 | 1260 | 1224 | -0.236 | 0.071 | 0.009 | 0.158 | 0.115 |
|---|
| UBE2D2 |   | 1 | 1195 | 1688 | 1703 | 0.212 | 0.002 | 0.217 | undef | 0.090 |
|---|
| CDH2 |   | 10 | 167 | 727 | 702 | 0.169 | 0.109 | -0.016 | 0.123 | 0.155 |
|---|
| SFN |   | 25 | 59 | 478 | 454 | 0.141 | 0.270 | 0.023 | -0.102 | 0.101 |
|---|
GO Enrichment output for subnetwork 6763 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 7.126E-07 | 1.741E-03 |
|---|
| neural plate morphogenesis | GO:0001839 |  | 1.422E-06 | 1.737E-03 |
|---|
| neural plate development | GO:0001840 |  | 2.833E-06 | 2.307E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 3.365E-06 | 2.055E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.935E-06 | 1.923E-03 |
|---|
| male meiosis | GO:0007140 |  | 7.871E-06 | 3.205E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 7.871E-06 | 2.747E-03 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 8.607E-06 | 2.628E-03 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.176E-05 | 3.192E-03 |
|---|
| DNA damage response. signal transduction by p53 class mediator resulting in induction of apoptosis | GO:0042771 |  | 1.674E-05 | 4.088E-03 |
|---|
| protein sumoylation | GO:0016925 |  | 1.674E-05 | 3.717E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 8.535E-07 | 2.053E-03 |
|---|
| male meiosis | GO:0007140 |  | 3.249E-06 | 3.909E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 4.026E-06 | 3.228E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.895E-06 | 2.945E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 9.024E-06 | 4.342E-03 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 1.029E-05 | 4.125E-03 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.348E-05 | 4.634E-03 |
|---|
| DNA damage response. signal transduction by p53 class mediator resulting in induction of apoptosis | GO:0042771 |  | 1.918E-05 | 5.769E-03 |
|---|
| protein sumoylation | GO:0016925 |  | 1.918E-05 | 5.128E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 1.99E-05 | 4.788E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 3.509E-05 | 7.676E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.844E-06 | 6.541E-03 |
|---|
| male meiosis | GO:0007140 |  | 8.09E-06 | 9.304E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 8.494E-06 | 6.512E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.167E-05 | 6.708E-03 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.24E-05 | 0.01030623 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.24E-05 | 8.589E-03 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 2.388E-05 | 7.845E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 4.543E-05 | 0.01305974 |
|---|
| response to X-ray | GO:0010165 |  | 4.749E-05 | 0.01213516 |
|---|
| DNA damage response. signal transduction by p53 class mediator resulting in induction of apoptosis | GO:0042771 |  | 4.749E-05 | 0.01092164 |
|---|
| protein sumoylation | GO:0016925 |  | 4.749E-05 | 9.929E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 5.33E-06 | 9.823E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.06E-05 | 9.765E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.162E-05 | 7.141E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.33E-05 | 6.129E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 1.722E-05 | 6.347E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.932E-05 | 9.007E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 4.715E-05 | 0.01241281 |
|---|
| synapse organization | GO:0050808 |  | 5.527E-05 | 0.01273238 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 6.21E-05 | 0.01271664 |
|---|
| positive regulation of transferase activity | GO:0051347 |  | 7.932E-05 | 0.0146185 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.454E-04 | 0.02436289 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.844E-06 | 6.541E-03 |
|---|
| male meiosis | GO:0007140 |  | 8.09E-06 | 9.304E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 8.494E-06 | 6.512E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.167E-05 | 6.708E-03 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.24E-05 | 0.01030623 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.24E-05 | 8.589E-03 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 2.388E-05 | 7.845E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 4.543E-05 | 0.01305974 |
|---|
| response to X-ray | GO:0010165 |  | 4.749E-05 | 0.01213516 |
|---|
| DNA damage response. signal transduction by p53 class mediator resulting in induction of apoptosis | GO:0042771 |  | 4.749E-05 | 0.01092164 |
|---|
| protein sumoylation | GO:0016925 |  | 4.749E-05 | 9.929E-03 |
|---|



