Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6614
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7536 | 3.529e-03 | 5.750e-04 | 7.712e-01 | 1.565e-06 |
|---|
| Loi | 0.2305 | 8.000e-02 | 1.129e-02 | 4.276e-01 | 3.862e-04 |
|---|
| Schmidt | 0.6739 | 0.000e+00 | 0.000e+00 | 4.435e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.471e-03 | 0.000e+00 |
|---|
| Wang | 0.2623 | 2.584e-03 | 4.295e-02 | 3.923e-01 | 4.354e-05 |
|---|
Expression data for subnetwork 6614 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
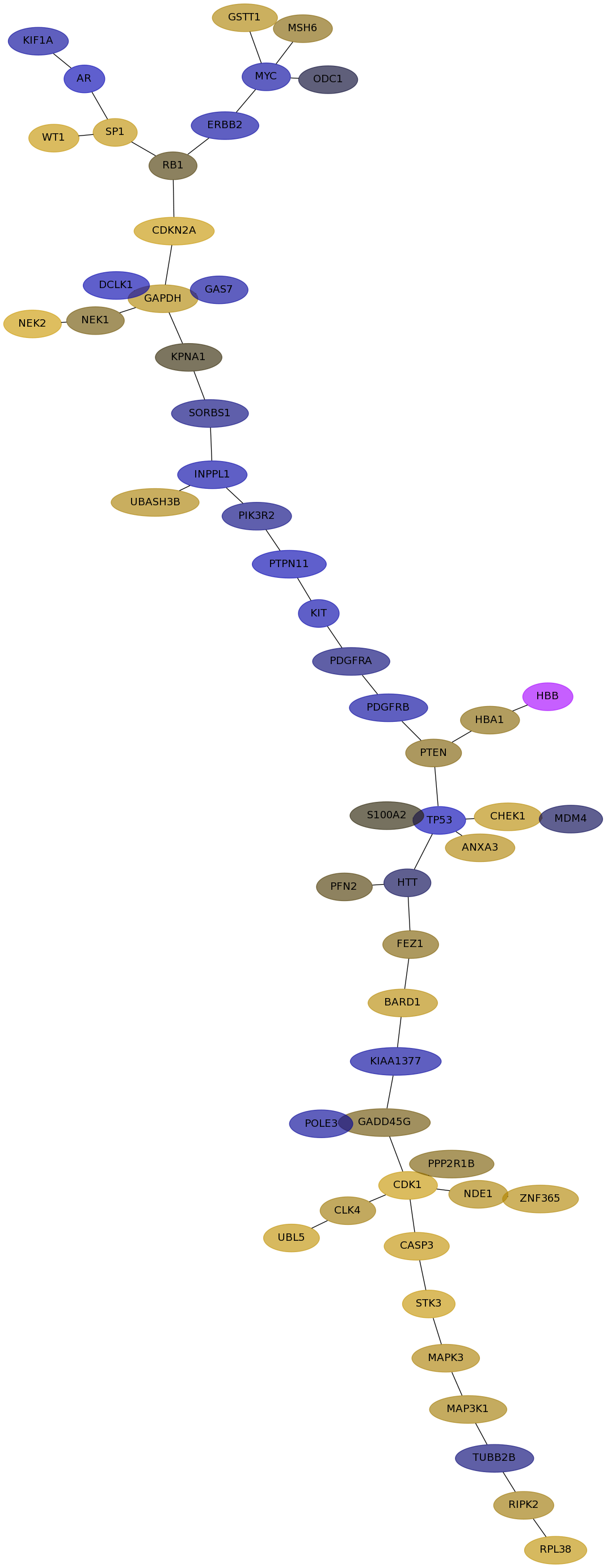
Score for each gene in subnetwork 6614 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| ANXA3 |   | 3 | 557 | 141 | 170 | 0.152 | 0.132 | 0.194 | 0.090 | 0.030 |
|---|
| CASP3 |   | 11 | 148 | 141 | 143 | 0.234 | -0.032 | -0.142 | -0.097 | -0.084 |
|---|
| INPPL1 |   | 6 | 301 | 141 | 152 | -0.122 | 0.072 | -0.087 | 0.174 | 0.156 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| GAS7 |   | 3 | 557 | 83 | 117 | -0.093 | -0.082 | 0.172 | 0.120 | 0.064 |
|---|
| UBASH3B |   | 1 | 1195 | 141 | 215 | 0.134 | 0.276 | undef | undef | undef |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| MDM4 |   | 9 | 196 | 141 | 147 | -0.016 | 0.004 | 0.164 | 0.107 | 0.105 |
|---|
| GSTT1 |   | 15 | 111 | 1 | 32 | 0.148 | 0.107 | -0.190 | 0.022 | -0.064 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CHEK1 |   | 18 | 86 | 141 | 136 | 0.189 | -0.134 | 0.119 | -0.088 | -0.054 |
|---|
| KIAA1377 |   | 18 | 86 | 141 | 136 | -0.095 | 0.116 | undef | 0.198 | undef |
|---|
| S100A2 |   | 6 | 301 | 1 | 52 | 0.005 | 0.047 | 0.137 | -0.029 | -0.008 |
|---|
| PFN2 |   | 11 | 148 | 141 | 143 | 0.014 | -0.001 | 0.286 | -0.002 | -0.056 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| PDGFRA |   | 10 | 167 | 141 | 146 | -0.035 | 0.008 | 0.118 | 0.107 | 0.119 |
|---|
| UBL5 |   | 1 | 1195 | 141 | 215 | 0.229 | 0.031 | 0.069 | undef | -0.094 |
|---|
| PPP2R1B |   | 6 | 301 | 141 | 152 | 0.042 | 0.002 | -0.002 | undef | 0.112 |
|---|
| HBB |   | 1 | 1195 | 141 | 215 | undef | -0.026 | 0.180 | 0.097 | 0.137 |
|---|
| RB1 |   | 31 | 43 | 1 | 22 | 0.013 | -0.051 | 0.057 | 0.100 | -0.082 |
|---|
| HBA1 |   | 3 | 557 | 141 | 170 | 0.058 | -0.042 | 0.140 | 0.142 | 0.131 |
|---|
| STK3 |   | 4 | 440 | 141 | 165 | 0.254 | 0.007 | 0.106 | 0.326 | 0.198 |
|---|
| PTEN |   | 27 | 50 | 141 | 135 | 0.045 | 0.055 | 0.017 | 0.183 | -0.041 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| PIK3R2 |   | 4 | 440 | 141 | 165 | -0.047 | 0.015 | -0.147 | undef | -0.135 |
|---|
| PTPN11 |   | 20 | 81 | 83 | 94 | -0.152 | 0.014 | 0.066 | 0.304 | 0.089 |
|---|
| NEK2 |   | 8 | 222 | 141 | 149 | 0.292 | -0.106 | 0.020 | 0.083 | 0.165 |
|---|
| CLK4 |   | 1 | 1195 | 141 | 215 | 0.105 | 0.032 | undef | 0.076 | undef |
|---|
| WT1 |   | 33 | 37 | 1 | 21 | 0.254 | -0.039 | 0.036 | undef | 0.172 |
|---|
| POLE3 |   | 17 | 95 | 141 | 138 | -0.083 | -0.033 | 0.231 | 0.096 | 0.011 |
|---|
| ZNF365 |   | 4 | 440 | 141 | 165 | 0.161 | 0.190 | -0.050 | -0.053 | -0.008 |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| MSH6 |   | 12 | 140 | 1 | 36 | 0.052 | -0.057 | 0.051 | 0.126 | 0.064 |
|---|
| PDGFRB |   | 12 | 140 | 141 | 142 | -0.094 | 0.224 | 0.127 | 0.153 | 0.189 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| MAP3K1 |   | 5 | 360 | 141 | 155 | 0.112 | 0.039 | undef | undef | undef |
|---|
| GAPDH |   | 11 | 148 | 141 | 143 | 0.159 | -0.040 | -0.053 | 0.088 | 0.029 |
|---|
| RIPK2 |   | 2 | 743 | 141 | 197 | 0.101 | -0.133 | -0.032 | -0.063 | 0.034 |
|---|
| RPL38 |   | 2 | 743 | 141 | 197 | 0.222 | 0.151 | 0.241 | -0.203 | -0.171 |
|---|
| FEZ1 |   | 8 | 222 | 141 | 149 | 0.047 | -0.020 | 0.154 | 0.165 | -0.128 |
|---|
| ODC1 |   | 4 | 440 | 141 | 165 | -0.006 | -0.099 | 0.209 | -0.207 | -0.107 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| MAPK3 |   | 6 | 301 | 141 | 152 | 0.137 | -0.102 | -0.126 | undef | -0.086 |
|---|
| KPNA1 |   | 9 | 196 | 141 | 147 | 0.007 | 0.152 | -0.083 | 0.073 | 0.086 |
|---|
| NEK1 |   | 15 | 111 | 141 | 139 | 0.032 | 0.184 | 0.049 | undef | 0.135 |
|---|
| KIF1A |   | 2 | 743 | 141 | 197 | -0.089 | -0.067 | 0.178 | 0.007 | -0.047 |
|---|
| HTT |   | 14 | 117 | 141 | 140 | -0.016 | -0.015 | 0.075 | 0.092 | -0.093 |
|---|
| DCLK1 |   | 7 | 256 | 141 | 151 | -0.145 | -0.006 | -0.048 | 0.188 | -0.035 |
|---|
| TUBB2B |   | 13 | 124 | 141 | 141 | -0.036 | 0.304 | 0.045 | 0.187 | 0.139 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| NDE1 |   | 3 | 557 | 141 | 170 | 0.142 | -0.058 | 0.142 | 0.107 | 0.111 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
GO Enrichment output for subnetwork 6614 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell cycle checkpoint | GO:0000075 |  | 9.978E-12 | 2.438E-08 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.775E-11 | 2.168E-08 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 3.832E-11 | 3.12E-08 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.274E-11 | 3.832E-08 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 7.369E-11 | 3.601E-08 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 1.189E-10 | 4.84E-08 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 1.263E-10 | 4.407E-08 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 1.37E-10 | 4.182E-08 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 3.857E-10 | 1.047E-07 |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 1.116E-09 | 2.727E-07 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 3.339E-09 | 7.415E-07 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell cycle checkpoint | GO:0000075 |  | 2.055E-11 | 4.943E-08 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 3.125E-11 | 3.759E-08 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 7.519E-11 | 6.03E-08 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 1.028E-10 | 6.183E-08 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.516E-10 | 7.295E-08 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 1.987E-10 | 7.967E-08 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 2.322E-10 | 7.98E-08 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 2.33E-10 | 7.006E-08 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 6.779E-10 | 1.812E-07 |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 1.757E-09 | 4.227E-07 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 5.252E-09 | 1.149E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 2.411E-11 | 5.546E-08 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 9.957E-10 | 1.145E-06 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 1.784E-09 | 1.368E-06 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 3.903E-09 | 2.244E-06 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 1.358E-08 | 6.248E-06 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 1.938E-08 | 7.43E-06 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 2.449E-08 | 8.047E-06 |
|---|
| organ growth | GO:0035265 |  | 2.71E-08 | 7.79E-06 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 2.71E-08 | 6.925E-06 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 3.272E-08 | 7.526E-06 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 5.397E-08 | 1.128E-05 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| oxygen transport | GO:0015671 |  | 3.751E-08 | 6.913E-05 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 1.339E-07 | 1.234E-04 |
|---|
| gas transport | GO:0015669 |  | 1.339E-07 | 8.225E-05 |
|---|
| meiosis | GO:0007126 |  | 2.214E-06 | 1.02E-03 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 2.214E-06 | 8.161E-04 |
|---|
| meiotic cell cycle | GO:0051321 |  | 2.538E-06 | 7.797E-04 |
|---|
| meiosis I | GO:0007127 |  | 3.128E-06 | 8.235E-04 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 3.74E-06 | 8.617E-04 |
|---|
| negative regulation of T cell proliferation | GO:0042130 |  | 3.881E-06 | 7.948E-04 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 3.945E-06 | 7.27E-04 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 6.039E-06 | 1.012E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 2.411E-11 | 5.546E-08 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 9.957E-10 | 1.145E-06 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 1.784E-09 | 1.368E-06 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 3.903E-09 | 2.244E-06 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 1.358E-08 | 6.248E-06 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 1.938E-08 | 7.43E-06 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 2.449E-08 | 8.047E-06 |
|---|
| organ growth | GO:0035265 |  | 2.71E-08 | 7.79E-06 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 2.71E-08 | 6.925E-06 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 3.272E-08 | 7.526E-06 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 5.397E-08 | 1.128E-05 |
|---|



