Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6581
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7541 | 3.498e-03 | 5.680e-04 | 7.700e-01 | 1.530e-06 |
|---|
| Loi | 0.2305 | 7.991e-02 | 1.127e-02 | 4.274e-01 | 3.847e-04 |
|---|
| Schmidt | 0.6735 | 0.000e+00 | 0.000e+00 | 4.484e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.475e-03 | 0.000e+00 |
|---|
| Wang | 0.2619 | 2.631e-03 | 4.338e-02 | 3.943e-01 | 4.501e-05 |
|---|
Expression data for subnetwork 6581 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
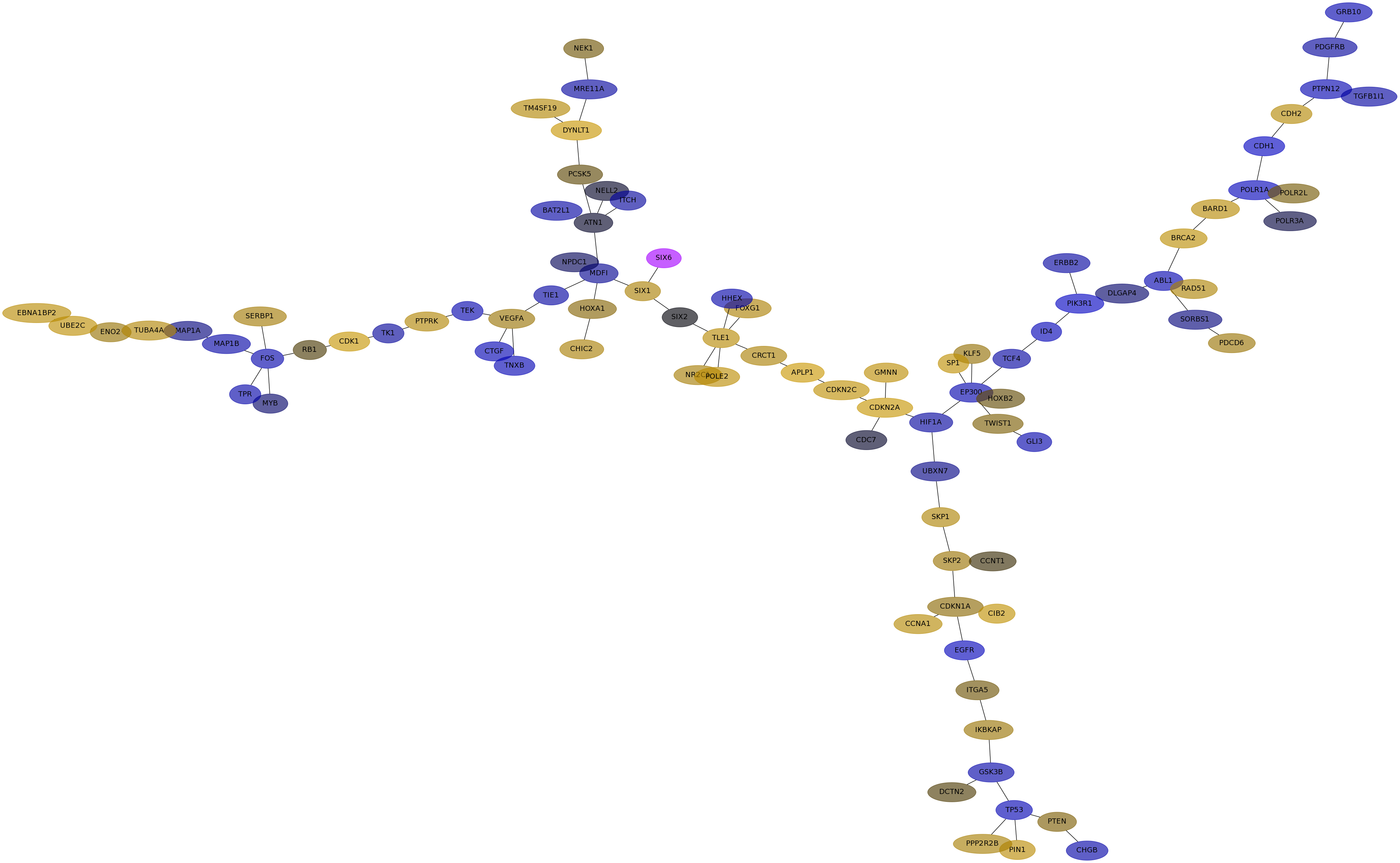
Score for each gene in subnetwork 6581 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| TEK |   | 18 | 86 | 83 | 95 | -0.149 | -0.049 | 0.174 | 0.015 | 0.113 |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| SIX6 |   | 2 | 743 | 727 | 734 | undef | -0.032 | 0.000 | 0.106 | 0.153 |
|---|
| POLR1A |   | 6 | 301 | 457 | 452 | -0.193 | -0.066 | undef | 0.093 | undef |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| CIB2 |   | 18 | 86 | 590 | 564 | 0.228 | -0.149 | 0.106 | 0.127 | 0.190 |
|---|
| TNXB |   | 4 | 440 | 727 | 723 | -0.169 | -0.076 | 0.160 | 0.182 | 0.173 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| ID4 |   | 35 | 34 | 236 | 223 | -0.179 | 0.137 | 0.241 | undef | 0.036 |
|---|
| EBNA1BP2 |   | 1 | 1195 | 727 | 771 | 0.186 | -0.041 | -0.003 | -0.061 | -0.058 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| TM4SF19 |   | 1 | 1195 | 727 | 771 | 0.163 | -0.182 | undef | undef | undef |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| CDKN2C |   | 16 | 104 | 236 | 227 | 0.216 | -0.106 | 0.202 | -0.024 | -0.081 |
|---|
| NELL2 |   | 7 | 256 | 727 | 714 | -0.005 | 0.138 | 0.421 | -0.027 | 0.074 |
|---|
| CTGF |   | 32 | 40 | 318 | 290 | -0.169 | 0.181 | 0.126 | 0.101 | 0.209 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| PIN1 |   | 24 | 62 | 179 | 181 | 0.194 | 0.040 | -0.040 | 0.065 | -0.048 |
|---|
| CRCT1 |   | 2 | 743 | 727 | 734 | 0.135 | 0.050 | 0.000 | -0.103 | 0.154 |
|---|
| MRE11A |   | 1 | 1195 | 727 | 771 | -0.098 | -0.093 | 0.105 | -0.006 | 0.065 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| BAT2L1 |   | 1 | 1195 | 727 | 771 | -0.111 | 0.240 | 0.093 | 0.189 | 0.086 |
|---|
| NR2C1 |   | 8 | 222 | 727 | 703 | 0.121 | 0.039 | 0.119 | -0.197 | -0.026 |
|---|
| PDGFRB |   | 12 | 140 | 141 | 142 | -0.094 | 0.224 | 0.127 | 0.153 | 0.189 |
|---|
| PTPRK |   | 14 | 117 | 83 | 96 | 0.156 | 0.141 | -0.094 | 0.048 | -0.024 |
|---|
| TUBA4A |   | 7 | 256 | 1 | 47 | 0.157 | 0.136 | -0.010 | undef | 0.156 |
|---|
| POLR2L |   | 5 | 360 | 727 | 722 | 0.037 | -0.012 | 0.224 | 0.054 | 0.015 |
|---|
| ITCH |   | 1 | 1195 | 727 | 771 | -0.097 | -0.150 | -0.170 | 0.006 | -0.120 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| HHEX |   | 4 | 440 | 727 | 723 | -0.130 | -0.178 | 0.177 | -0.180 | 0.005 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| CCNT1 |   | 6 | 301 | 478 | 473 | 0.008 | -0.028 | 0.162 | 0.007 | -0.213 |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| TLE1 |   | 4 | 440 | 727 | 723 | 0.183 | 0.050 | 0.030 | 0.080 | 0.055 |
|---|
| SKP1 |   | 2 | 743 | 727 | 734 | 0.143 | 0.037 | 0.055 | 0.148 | -0.110 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| GRB10 |   | 3 | 557 | 727 | 728 | -0.149 | 0.182 | -0.070 | -0.000 | 0.081 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| PCSK5 |   | 2 | 743 | 727 | 734 | 0.021 | 0.074 | 0.121 | undef | 0.092 |
|---|
| ENO2 |   | 3 | 557 | 727 | 728 | 0.085 | 0.114 | 0.148 | -0.071 | 0.208 |
|---|
| DCTN2 |   | 8 | 222 | 366 | 373 | 0.014 | 0.150 | 0.061 | 0.113 | 0.303 |
|---|
| DLGAP4 |   | 7 | 256 | 727 | 714 | -0.029 | 0.180 | -0.201 | 0.135 | -0.043 |
|---|
| CHIC2 |   | 2 | 743 | 727 | 734 | 0.128 | -0.154 | 0.120 | 0.154 | -0.008 |
|---|
| IKBKAP |   | 10 | 167 | 412 | 408 | 0.089 | 0.184 | 0.142 | 0.105 | -0.017 |
|---|
| FOXG1 |   | 6 | 301 | 727 | 716 | 0.136 | -0.156 | -0.126 | -0.013 | -0.068 |
|---|
| HOXB2 |   | 33 | 37 | 318 | 289 | 0.024 | -0.143 | 0.076 | 0.247 | -0.184 |
|---|
| GMNN |   | 2 | 743 | 1 | 66 | 0.201 | -0.071 | 0.104 | 0.066 | 0.158 |
|---|
| UBXN7 |   | 3 | 557 | 727 | 728 | -0.055 | 0.153 | 0.224 | 0.167 | 0.175 |
|---|
| ABL1 |   | 13 | 124 | 727 | 699 | -0.152 | 0.200 | 0.108 | undef | -0.059 |
|---|
| RB1 |   | 31 | 43 | 1 | 22 | 0.013 | -0.051 | 0.057 | 0.100 | -0.082 |
|---|
| APLP1 |   | 8 | 222 | 727 | 703 | 0.284 | 0.043 | -0.105 | -0.121 | -0.190 |
|---|
| SIX1 |   | 2 | 743 | 727 | 734 | 0.133 | -0.134 | 0.005 | -0.084 | 0.068 |
|---|
| GLI3 |   | 21 | 73 | 478 | 456 | -0.138 | 0.080 | -0.007 | 0.282 | -0.018 |
|---|
| MAP1A |   | 12 | 140 | 1 | 36 | -0.045 | 0.143 | -0.054 | 0.162 | 0.147 |
|---|
| TCF4 |   | 2 | 743 | 727 | 734 | -0.109 | 0.229 | 0.063 | 0.025 | 0.097 |
|---|
| PTEN |   | 27 | 50 | 141 | 135 | 0.045 | 0.055 | 0.017 | 0.183 | -0.041 |
|---|
| CDC7 |   | 1 | 1195 | 727 | 771 | -0.005 | -0.168 | 0.025 | undef | 0.126 |
|---|
| SERBP1 |   | 2 | 743 | 727 | 734 | 0.111 | 0.022 | 0.041 | -0.093 | 0.019 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| MYB |   | 28 | 47 | 366 | 344 | -0.028 | 0.075 | 0.002 | 0.065 | -0.012 |
|---|
| MDFI |   | 17 | 95 | 236 | 226 | -0.075 | -0.001 | 0.177 | 0.169 | -0.112 |
|---|
| CHGB |   | 7 | 256 | 682 | 664 | -0.126 | -0.041 | 0.018 | 0.102 | -0.131 |
|---|
| TK1 |   | 4 | 440 | 727 | 723 | -0.089 | -0.156 | 0.029 | 0.119 | -0.052 |
|---|
| DYNLT1 |   | 2 | 743 | 727 | 734 | 0.275 | -0.041 | -0.176 | -0.004 | -0.168 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| SKP2 |   | 29 | 45 | 1 | 23 | 0.092 | -0.043 | 0.237 | undef | -0.009 |
|---|
| PTPN12 |   | 6 | 301 | 478 | 473 | -0.159 | 0.153 | 0.031 | 0.083 | 0.042 |
|---|
| ITGA5 |   | 26 | 52 | 179 | 177 | 0.031 | 0.192 | 0.125 | 0.133 | 0.202 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| NPDC1 |   | 3 | 557 | 727 | 728 | -0.019 | 0.221 | 0.044 | 0.006 | -0.146 |
|---|
| HOXA1 |   | 1 | 1195 | 727 | 771 | 0.055 | -0.047 | 0.147 | undef | 0.042 |
|---|
| POLR3A |   | 8 | 222 | 296 | 287 | -0.010 | -0.032 | undef | 0.091 | undef |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| TPR |   | 11 | 148 | 727 | 701 | -0.130 | 0.120 | -0.036 | -0.085 | 0.156 |
|---|
| PDCD6 |   | 10 | 167 | 1 | 40 | 0.091 | -0.060 | undef | undef | undef |
|---|
| NEK1 |   | 15 | 111 | 141 | 139 | 0.032 | 0.184 | 0.049 | undef | 0.135 |
|---|
| POLE2 |   | 4 | 440 | 727 | 723 | 0.200 | -0.203 | 0.049 | 0.000 | -0.010 |
|---|
| HIF1A |   | 13 | 124 | 236 | 230 | -0.098 | 0.224 | -0.118 | 0.135 | 0.356 |
|---|
| RAD51 |   | 13 | 124 | 727 | 699 | 0.146 | -0.098 | 0.135 | -0.070 | 0.061 |
|---|
| ATN1 |   | 7 | 256 | 412 | 414 | -0.005 | -0.093 | 0.005 | -0.185 | -0.136 |
|---|
| TIE1 |   | 18 | 86 | 366 | 357 | -0.108 | 0.005 | 0.141 | -0.108 | 0.252 |
|---|
| UBE2C |   | 1 | 1195 | 727 | 771 | 0.210 | -0.211 | 0.045 | undef | 0.086 |
|---|
| CDH2 |   | 10 | 167 | 727 | 702 | 0.169 | 0.109 | -0.016 | 0.123 | 0.155 |
|---|
| SIX2 |   | 2 | 743 | 727 | 734 | -0.001 | -0.092 | -0.196 | 0.161 | 0.137 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
GO Enrichment output for subnetwork 6581 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| elastic fiber assembly | GO:0048251 |  | 6.741E-17 | 1.647E-13 |
|---|
| collagen metabolic process | GO:0032963 |  | 2.184E-13 | 2.667E-10 |
|---|
| multicellular organismal macromolecule metabolic process | GO:0044259 |  | 5.411E-13 | 4.407E-10 |
|---|
| multicellular organismal metabolic process | GO:0044236 |  | 2.603E-12 | 1.59E-09 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.974E-12 | 3.896E-09 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.15E-11 | 2.504E-08 |
|---|
| aging | GO:0007568 |  | 7.241E-11 | 2.527E-08 |
|---|
| extracellular matrix organization | GO:0030198 |  | 3.181E-10 | 9.715E-08 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 3.625E-10 | 9.839E-08 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.475E-10 | 1.093E-07 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 5.199E-10 | 1.155E-07 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| elastic fiber assembly | GO:0048251 |  | 1.524E-14 | 3.668E-11 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.488E-11 | 1.79E-08 |
|---|
| collagen metabolic process | GO:0032963 |  | 1.857E-11 | 1.489E-08 |
|---|
| multicellular organismal macromolecule metabolic process | GO:0044259 |  | 4.203E-11 | 2.528E-08 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.143E-10 | 5.5E-08 |
|---|
| multicellular organismal metabolic process | GO:0044236 |  | 1.722E-10 | 6.906E-08 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.44E-10 | 2.214E-07 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 7.955E-10 | 2.393E-07 |
|---|
| interphase | GO:0051325 |  | 8.777E-10 | 2.346E-07 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 4.839E-09 | 1.164E-06 |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 8.478E-09 | 1.854E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| elastic fiber assembly | GO:0048251 |  | 1.461E-13 | 3.359E-10 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.311E-10 | 1.507E-07 |
|---|
| collagen metabolic process | GO:0032963 |  | 1.75E-10 | 1.342E-07 |
|---|
| multicellular organismal macromolecule metabolic process | GO:0044259 |  | 4.059E-10 | 2.334E-07 |
|---|
| multicellular organismal metabolic process | GO:0044236 |  | 1.718E-09 | 7.905E-07 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.046E-09 | 1.934E-06 |
|---|
| interphase | GO:0051325 |  | 6.335E-09 | 2.081E-06 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 1.332E-08 | 3.829E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.624E-08 | 4.149E-06 |
|---|
| extracellular structure organization | GO:0043062 |  | 2.01E-08 | 4.622E-06 |
|---|
| extracellular matrix organization | GO:0030198 |  | 6.96E-08 | 1.455E-05 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 8.65E-08 | 1.594E-04 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 1.425E-07 | 1.313E-04 |
|---|
| regulation of cell migration | GO:0030334 |  | 2.716E-07 | 1.668E-04 |
|---|
| skeletal system morphogenesis | GO:0048705 |  | 2.795E-07 | 1.288E-04 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 3.441E-07 | 1.268E-04 |
|---|
| embryonic skeletal system morphogenesis | GO:0048704 |  | 3.651E-07 | 1.121E-04 |
|---|
| regulation of angiogenesis | GO:0045765 |  | 6.147E-07 | 1.618E-04 |
|---|
| regulation of locomotion | GO:0040012 |  | 6.275E-07 | 1.446E-04 |
|---|
| regulation of cell motion | GO:0051270 |  | 6.275E-07 | 1.285E-04 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 8.826E-07 | 1.627E-04 |
|---|
| embryonic skeletal system development | GO:0048706 |  | 1.498E-06 | 2.51E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| elastic fiber assembly | GO:0048251 |  | 1.461E-13 | 3.359E-10 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.311E-10 | 1.507E-07 |
|---|
| collagen metabolic process | GO:0032963 |  | 1.75E-10 | 1.342E-07 |
|---|
| multicellular organismal macromolecule metabolic process | GO:0044259 |  | 4.059E-10 | 2.334E-07 |
|---|
| multicellular organismal metabolic process | GO:0044236 |  | 1.718E-09 | 7.905E-07 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.046E-09 | 1.934E-06 |
|---|
| interphase | GO:0051325 |  | 6.335E-09 | 2.081E-06 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 1.332E-08 | 3.829E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.624E-08 | 4.149E-06 |
|---|
| extracellular structure organization | GO:0043062 |  | 2.01E-08 | 4.622E-06 |
|---|
| extracellular matrix organization | GO:0030198 |  | 6.96E-08 | 1.455E-05 |
|---|



