Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6578
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7543 | 3.490e-03 | 5.670e-04 | 7.696e-01 | 1.523e-06 |
|---|
| Loi | 0.2305 | 8.000e-02 | 1.129e-02 | 4.276e-01 | 3.861e-04 |
|---|
| Schmidt | 0.6734 | 0.000e+00 | 0.000e+00 | 4.494e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.474e-03 | 0.000e+00 |
|---|
| Wang | 0.2619 | 2.633e-03 | 4.340e-02 | 3.944e-01 | 4.507e-05 |
|---|
Expression data for subnetwork 6578 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
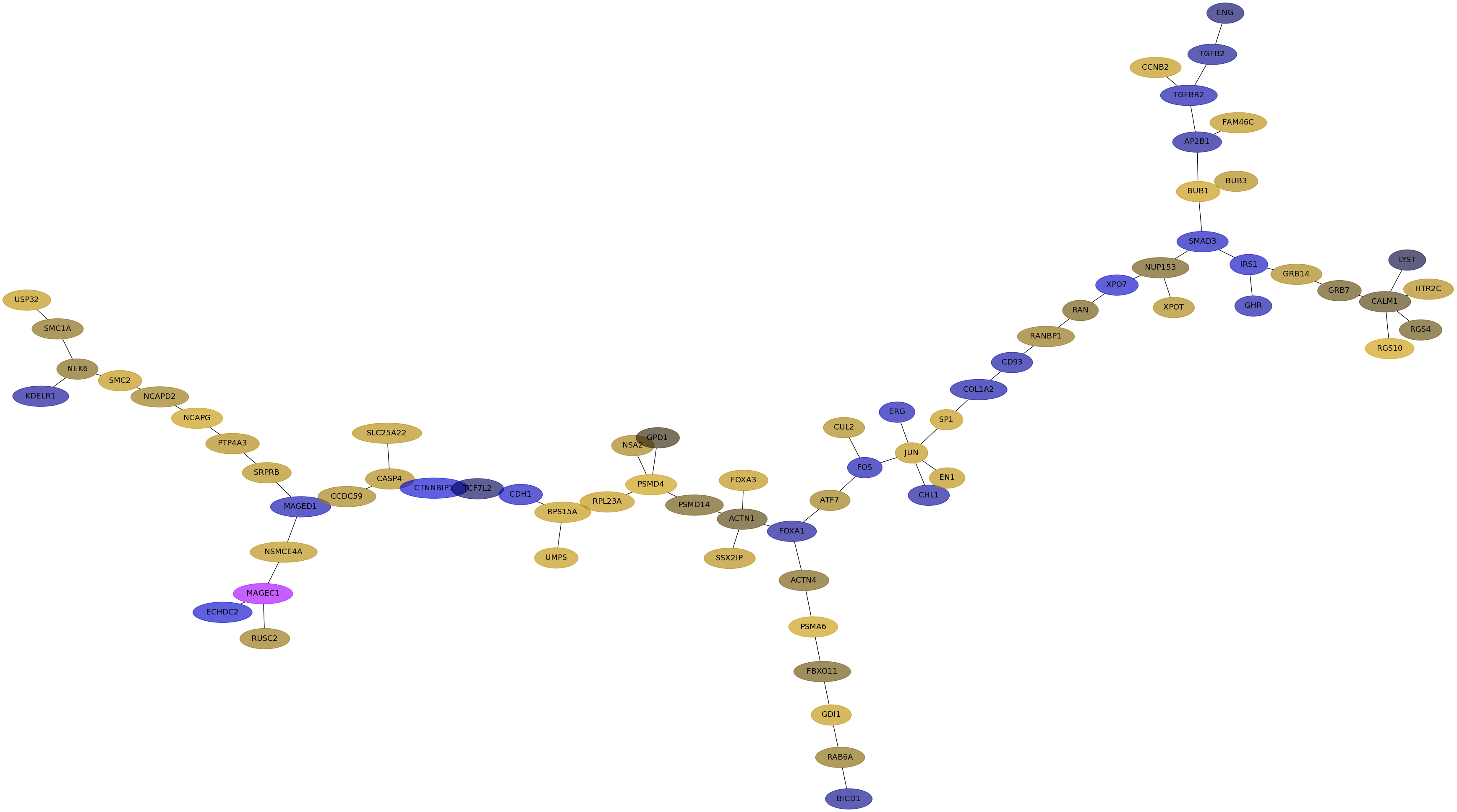
Subnetwork structure for each dataset
- IPC
Score for each gene in subnetwork 6578 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| RGS4 |   | 4 | 440 | 842 | 831 | 0.022 | 0.160 | -0.006 | undef | 0.371 |
|---|
| MAGED1 |   | 5 | 360 | 842 | 828 | -0.141 | 0.284 | -0.152 | 0.240 | 0.045 |
|---|
| PTP4A3 |   | 9 | 196 | 412 | 411 | 0.137 | -0.196 | 0.085 | undef | -0.004 |
|---|
| FAM46C |   | 2 | 743 | 842 | 854 | 0.183 | -0.142 | -0.141 | -0.090 | -0.127 |
|---|
| RPL23A |   | 5 | 360 | 842 | 828 | 0.208 | -0.007 | 0.024 | -0.242 | 0.055 |
|---|
| ENG |   | 2 | 743 | 842 | 854 | -0.027 | 0.062 | 0.130 | 0.174 | 0.010 |
|---|
| NCAPD2 |   | 1 | 1195 | 842 | 877 | 0.080 | -0.105 | 0.006 | -0.078 | -0.138 |
|---|
| SRPRB |   | 1 | 1195 | 842 | 877 | 0.148 | -0.006 | -0.124 | -0.048 | 0.019 |
|---|
| PSMA6 |   | 5 | 360 | 842 | 828 | 0.280 | -0.118 | -0.018 | undef | -0.091 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| NEK6 |   | 3 | 557 | 842 | 834 | 0.041 | -0.049 | undef | 0.271 | undef |
|---|
| PSMD14 |   | 4 | 440 | 842 | 831 | 0.027 | 0.135 | -0.179 | -0.018 | -0.031 |
|---|
| AP2B1 |   | 6 | 301 | 780 | 764 | -0.070 | -0.106 | 0.070 | 0.252 | 0.094 |
|---|
| NUP153 |   | 1 | 1195 | 842 | 877 | 0.026 | -0.038 | 0.125 | -0.102 | 0.141 |
|---|
| CHL1 |   | 2 | 743 | 842 | 854 | -0.088 | 0.161 | 0.202 | 0.103 | -0.105 |
|---|
| RAN |   | 3 | 557 | 236 | 265 | 0.030 | -0.006 | 0.062 | 0.016 | undef |
|---|
| LYST |   | 1 | 1195 | 842 | 877 | -0.008 | 0.170 | -0.172 | 0.296 | -0.059 |
|---|
| SLC25A22 |   | 1 | 1195 | 842 | 877 | 0.167 | -0.080 | -0.071 | undef | -0.079 |
|---|
| ACTN1 |   | 18 | 86 | 318 | 313 | 0.016 | 0.347 | 0.068 | 0.100 | 0.314 |
|---|
| SMC2 |   | 4 | 440 | 83 | 112 | 0.202 | -0.108 | 0.119 | 0.132 | 0.079 |
|---|
| CASP4 |   | 1 | 1195 | 842 | 877 | 0.134 | -0.070 | -0.068 | -0.227 | -0.016 |
|---|
| XPOT |   | 1 | 1195 | 842 | 877 | 0.126 | -0.014 | -0.044 | -0.035 | 0.134 |
|---|
| NSA2 |   | 4 | 440 | 806 | 790 | 0.103 | 0.097 | 0.132 | -0.026 | 0.006 |
|---|
| SMAD3 |   | 3 | 557 | 842 | 834 | -0.176 | 0.063 | 0.239 | -0.068 | -0.012 |
|---|
| ECHDC2 |   | 2 | 743 | 842 | 854 | -0.315 | 0.242 | -0.069 | 0.051 | 0.090 |
|---|
| RGS10 |   | 2 | 743 | 842 | 854 | 0.295 | -0.214 | -0.002 | undef | -0.095 |
|---|
| SMC1A |   | 1 | 1195 | 842 | 877 | 0.048 | -0.088 | -0.012 | 0.184 | 0.070 |
|---|
| ACTN4 |   | 14 | 117 | 318 | 315 | 0.035 | 0.180 | undef | -0.048 | undef |
|---|
| MAGEC1 |   | 2 | 743 | 842 | 854 | undef | 0.029 | 0.168 | 0.225 | -0.132 |
|---|
| CCDC59 |   | 2 | 743 | 842 | 854 | 0.102 | -0.117 | 0.124 | -0.093 | 0.007 |
|---|
| CD93 |   | 2 | 743 | 842 | 854 | -0.102 | 0.036 | 0.160 | -0.146 | 0.254 |
|---|
| NSMCE4A |   | 2 | 743 | 842 | 854 | 0.177 | -0.116 | 0.290 | -0.022 | -0.091 |
|---|
| HTR2C |   | 2 | 743 | 842 | 854 | 0.135 | 0.027 | 0.162 | 0.138 | undef |
|---|
| GRB14 |   | 24 | 62 | 179 | 181 | 0.113 | -0.129 | 0.143 | -0.098 | 0.138 |
|---|
| RUSC2 |   | 1 | 1195 | 842 | 877 | 0.073 | -0.080 | 0.199 | -0.106 | 0.267 |
|---|
| ATF7 |   | 28 | 47 | 179 | 175 | 0.085 | 0.115 | -0.088 | 0.172 | -0.092 |
|---|
| FOXA3 |   | 12 | 140 | 318 | 322 | 0.192 | 0.289 | undef | -0.171 | undef |
|---|
| RAB6A |   | 2 | 743 | 842 | 854 | 0.054 | -0.037 | -0.038 | 0.208 | -0.164 |
|---|
| TGFB2 |   | 7 | 256 | 525 | 515 | -0.063 | 0.244 | 0.226 | 0.079 | 0.015 |
|---|
| XPO7 |   | 1 | 1195 | 842 | 877 | -0.245 | -0.039 | 0.142 | undef | -0.120 |
|---|
| TGFBR2 |   | 7 | 256 | 525 | 515 | -0.115 | -0.028 | 0.136 | 0.105 | -0.190 |
|---|
| NCAPG |   | 3 | 557 | 83 | 117 | 0.253 | -0.183 | 0.065 | undef | 0.020 |
|---|
| SSX2IP |   | 1 | 1195 | 842 | 877 | 0.160 | -0.068 | 0.015 | -0.065 | 0.084 |
|---|
| GPD1 |   | 7 | 256 | 658 | 644 | 0.006 | -0.019 | 0.112 | -0.034 | 0.033 |
|---|
| GDI1 |   | 3 | 557 | 842 | 834 | 0.219 | 0.094 | 0.049 | 0.181 | 0.049 |
|---|
| JUN |   | 26 | 52 | 179 | 177 | 0.200 | 0.051 | 0.220 | undef | 0.200 |
|---|
| CTNNBIP1 |   | 1 | 1195 | 842 | 877 | -0.342 | 0.068 | -0.049 | 0.101 | 0.163 |
|---|
| EN1 |   | 10 | 167 | 179 | 194 | 0.196 | 0.024 | 0.011 | -0.016 | 0.102 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| RANBP1 |   | 2 | 743 | 842 | 854 | 0.063 | -0.066 | 0.199 | -0.107 | -0.048 |
|---|
| BUB1 |   | 6 | 301 | 590 | 582 | 0.246 | -0.172 | 0.013 | -0.007 | 0.170 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| KDELR1 |   | 2 | 743 | 842 | 854 | -0.073 | -0.029 | -0.123 | 0.288 | 0.004 |
|---|
| ERG |   | 9 | 196 | 179 | 195 | -0.134 | -0.026 | 0.235 | -0.027 | 0.182 |
|---|
| BUB3 |   | 5 | 360 | 478 | 476 | 0.133 | -0.053 | 0.165 | 0.195 | 0.181 |
|---|
| CALM1 |   | 6 | 301 | 614 | 602 | 0.014 | 0.021 | 0.192 | 0.221 | 0.247 |
|---|
| PSMD4 |   | 11 | 148 | 296 | 282 | 0.285 | -0.243 | -0.043 | 0.087 | -0.045 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| TCF7L2 |   | 17 | 95 | 366 | 358 | -0.021 | 0.143 | 0.280 | undef | 0.101 |
|---|
| IRS1 |   | 9 | 196 | 842 | 805 | -0.219 | 0.065 | 0.165 | 0.102 | 0.064 |
|---|
| FBXO11 |   | 5 | 360 | 457 | 455 | 0.027 | 0.186 | -0.069 | 0.238 | 0.149 |
|---|
| BICD1 |   | 1 | 1195 | 842 | 877 | -0.059 | 0.045 | 0.142 | 0.147 | -0.148 |
|---|
| RPS15A |   | 2 | 743 | 842 | 854 | 0.221 | -0.019 | 0.189 | -0.123 | -0.060 |
|---|
| CUL2 |   | 7 | 256 | 842 | 816 | 0.124 | -0.192 | -0.090 | -0.035 | 0.166 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| USP32 |   | 2 | 743 | 842 | 854 | 0.204 | 0.263 | 0.137 | undef | 0.062 |
|---|
| UMPS |   | 3 | 557 | 842 | 834 | 0.229 | 0.047 | 0.080 | -0.098 | 0.021 |
|---|
| COL1A2 |   | 7 | 256 | 552 | 542 | -0.110 | 0.207 | 0.046 | 0.235 | 0.097 |
|---|
| CCNB2 |   | 4 | 440 | 842 | 831 | 0.206 | -0.119 | 0.004 | -0.006 | 0.134 |
|---|
| FOXA1 |   | 27 | 50 | 179 | 176 | -0.072 | 0.071 | -0.015 | 0.214 | 0.005 |
|---|
GO Enrichment output for subnetwork 6578 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mitotic sister chromatid segregation | GO:0000070 |  | 3.561E-08 | 8.698E-05 |
|---|
| sister chromatid segregation | GO:0000819 |  | 4.231E-08 | 5.168E-05 |
|---|
| chromosome segregation | GO:0007059 |  | 7.141E-08 | 5.815E-05 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 7.141E-08 | 4.362E-05 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 7.925E-08 | 3.872E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 7.925E-08 | 3.227E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 1.183E-07 | 4.128E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 2.256E-07 | 6.889E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 3.447E-07 | 9.356E-05 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 6.925E-07 | 1.692E-04 |
|---|
| spindle organization | GO:0007051 |  | 7.225E-07 | 1.605E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mitotic sister chromatid segregation | GO:0000070 |  | 6.158E-08 | 1.482E-04 |
|---|
| sister chromatid segregation | GO:0000819 |  | 7.314E-08 | 8.799E-05 |
|---|
| chromosome segregation | GO:0007059 |  | 1.337E-07 | 1.072E-04 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 1.337E-07 | 8.039E-05 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 1.483E-07 | 7.134E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.483E-07 | 5.945E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 2.209E-07 | 7.593E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 4.2E-07 | 1.263E-04 |
|---|
| regulation of ligase activity | GO:0051340 |  | 6.404E-07 | 1.712E-04 |
|---|
| spindle organization | GO:0007051 |  | 1.141E-06 | 2.745E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.282E-06 | 2.803E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mitotic sister chromatid segregation | GO:0000070 |  | 9.665E-08 | 2.223E-04 |
|---|
| sister chromatid segregation | GO:0000819 |  | 1.174E-07 | 1.351E-04 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 2.403E-07 | 1.842E-04 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 2.698E-07 | 1.552E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.698E-07 | 1.241E-04 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 4.204E-07 | 1.612E-04 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 7.749E-07 | 2.546E-04 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.129E-06 | 3.247E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.754E-06 | 4.484E-04 |
|---|
| spindle organization | GO:0007051 |  | 2.215E-06 | 5.094E-04 |
|---|
| chromosome segregation | GO:0007059 |  | 3.341E-06 | 6.985E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| chromosome segregation | GO:0007059 |  | 3.181E-07 | 5.863E-04 |
|---|
| mitotic sister chromatid segregation | GO:0000070 |  | 3.763E-07 | 3.468E-04 |
|---|
| sister chromatid segregation | GO:0000819 |  | 4.842E-07 | 2.975E-04 |
|---|
| spindle organization | GO:0007051 |  | 4.842E-07 | 2.231E-04 |
|---|
| regulation of mitotic metaphase/anaphase transition | GO:0030071 |  | 1.374E-06 | 5.064E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 4.03E-06 | 1.238E-03 |
|---|
| positive regulation of transcription factor import into nucleus | GO:0042993 |  | 8.017E-06 | 2.111E-03 |
|---|
| regulation of mitotic cell cycle | GO:0007346 |  | 1.156E-05 | 2.662E-03 |
|---|
| SMAD protein nuclear translocation | GO:0007184 |  | 1.396E-05 | 2.858E-03 |
|---|
| transmembrane receptor protein serine/threonine kinase signaling pathway | GO:0007178 |  | 2.445E-05 | 4.507E-03 |
|---|
| regulation of cell cycle process | GO:0010564 |  | 2.714E-05 | 4.548E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mitotic sister chromatid segregation | GO:0000070 |  | 9.665E-08 | 2.223E-04 |
|---|
| sister chromatid segregation | GO:0000819 |  | 1.174E-07 | 1.351E-04 |
|---|
| negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051436 |  | 2.403E-07 | 1.842E-04 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 2.698E-07 | 1.552E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.698E-07 | 1.241E-04 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 4.204E-07 | 1.612E-04 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 7.749E-07 | 2.546E-04 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.129E-06 | 3.247E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 1.754E-06 | 4.484E-04 |
|---|
| spindle organization | GO:0007051 |  | 2.215E-06 | 5.094E-04 |
|---|
| chromosome segregation | GO:0007059 |  | 3.341E-06 | 6.985E-04 |
|---|



