Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6558
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7542 | 3.494e-03 | 5.670e-04 | 7.698e-01 | 1.525e-06 |
|---|
| Loi | 0.2307 | 7.962e-02 | 1.119e-02 | 4.266e-01 | 3.801e-04 |
|---|
| Schmidt | 0.6729 | 0.000e+00 | 0.000e+00 | 4.548e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.459e-03 | 0.000e+00 |
|---|
| Wang | 0.2618 | 2.654e-03 | 4.359e-02 | 3.953e-01 | 4.574e-05 |
|---|
Expression data for subnetwork 6558 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
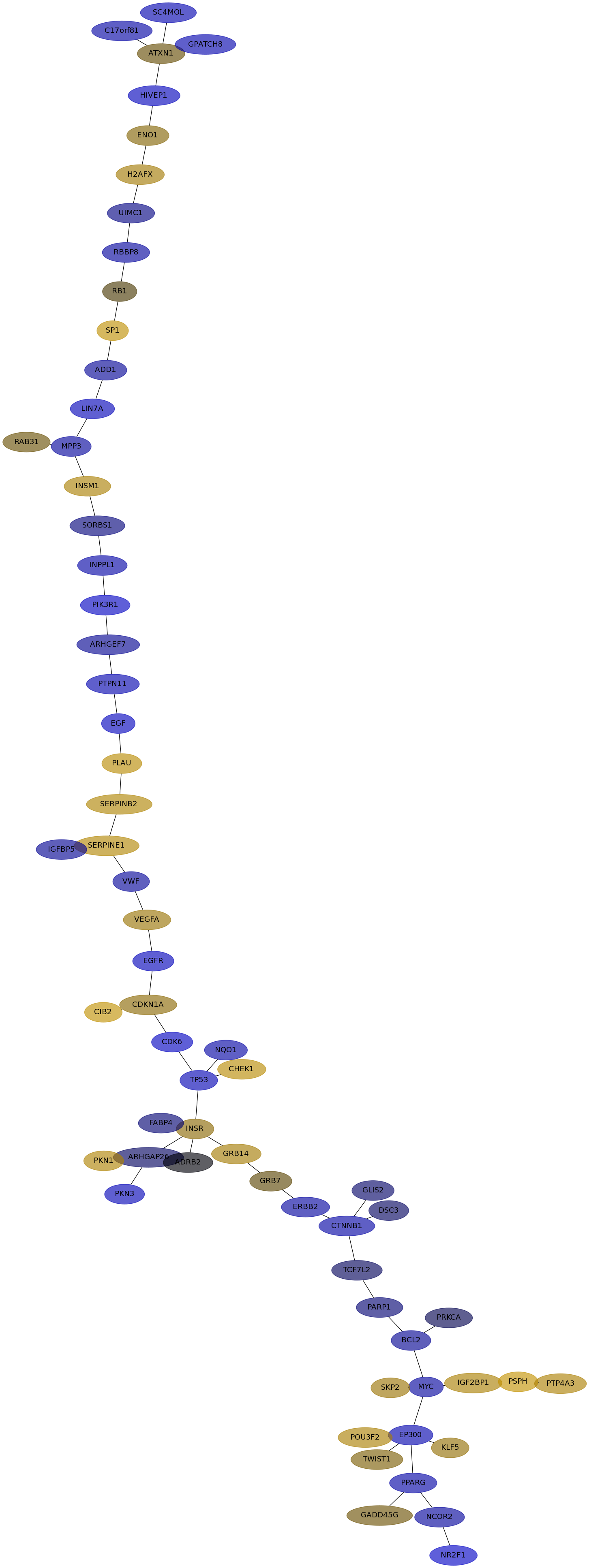
Score for each gene in subnetwork 6558 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| ARHGAP26 |   | 21 | 73 | 457 | 438 | -0.023 | 0.171 | -0.222 | 0.015 | -0.348 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| ADD1 |   | 8 | 222 | 366 | 373 | -0.085 | 0.116 | 0.035 | undef | -0.098 |
|---|
| NQO1 |   | 21 | 73 | 366 | 354 | -0.115 | 0.069 | -0.189 | 0.321 | 0.118 |
|---|
| EGF |   | 12 | 140 | 638 | 620 | -0.212 | 0.182 | -0.276 | -0.060 | 0.005 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| CIB2 |   | 18 | 86 | 590 | 564 | 0.228 | -0.149 | 0.106 | 0.127 | 0.190 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| PTP4A3 |   | 9 | 196 | 412 | 411 | 0.137 | -0.196 | 0.085 | undef | -0.004 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| PLAU |   | 12 | 140 | 179 | 192 | 0.187 | 0.153 | 0.141 | 0.072 | 0.192 |
|---|
| GLIS2 |   | 1 | 1195 | 1674 | 1690 | -0.027 | 0.190 | undef | undef | undef |
|---|
| LIN7A |   | 6 | 301 | 366 | 377 | -0.189 | -0.074 | 0.108 | 0.065 | -0.146 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| ENO1 |   | 3 | 557 | 1437 | 1418 | 0.054 | 0.211 | 0.010 | 0.017 | 0.178 |
|---|
| CDK6 |   | 16 | 104 | 318 | 314 | -0.229 | 0.107 | -0.028 | -0.118 | 0.104 |
|---|
| DSC3 |   | 1 | 1195 | 1674 | 1690 | -0.023 | 0.014 | 0.324 | -0.201 | 0.147 |
|---|
| PARP1 |   | 29 | 45 | 83 | 84 | -0.036 | -0.162 | -0.089 | 0.238 | 0.012 |
|---|
| PPARG |   | 7 | 256 | 179 | 201 | -0.119 | 0.011 | 0.105 | 0.059 | 0.039 |
|---|
| ATXN1 |   | 17 | 95 | 1 | 29 | 0.026 | 0.192 | 0.175 | 0.125 | 0.163 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| RBBP8 |   | 1 | 1195 | 1674 | 1690 | -0.098 | 0.013 | 0.127 | -0.115 | 0.070 |
|---|
| H2AFX |   | 7 | 256 | 179 | 201 | 0.111 | -0.072 | 0.121 | 0.165 | -0.010 |
|---|
| UIMC1 |   | 6 | 301 | 179 | 205 | -0.054 | 0.226 | 0.031 | -0.005 | 0.189 |
|---|
| SERPINE1 |   | 8 | 222 | 179 | 200 | 0.160 | 0.200 | 0.056 | 0.070 | 0.030 |
|---|
| MPP3 |   | 4 | 440 | 1650 | 1612 | -0.095 | -0.007 | -0.098 | 0.087 | -0.046 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| PSPH |   | 20 | 81 | 366 | 356 | 0.240 | -0.019 | -0.102 | 0.358 | 0.025 |
|---|
| HIVEP1 |   | 6 | 301 | 764 | 758 | -0.199 | 0.069 | 0.000 | 0.193 | -0.042 |
|---|
| FABP4 |   | 6 | 301 | 236 | 239 | -0.036 | -0.028 | 0.227 | 0.072 | 0.107 |
|---|
| POU3F2 |   | 17 | 95 | 928 | 891 | 0.135 | 0.114 | 0.155 | 0.253 | 0.040 |
|---|
| NCOR2 |   | 32 | 40 | 318 | 290 | -0.096 | 0.211 | -0.163 | 0.158 | 0.114 |
|---|
| INPPL1 |   | 6 | 301 | 141 | 152 | -0.122 | 0.072 | -0.087 | 0.174 | 0.156 |
|---|
| GRB14 |   | 24 | 62 | 179 | 181 | 0.113 | -0.129 | 0.143 | -0.098 | 0.138 |
|---|
| ARHGEF7 |   | 25 | 59 | 83 | 92 | -0.074 | -0.061 | 0.208 | 0.028 | 0.015 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| CHEK1 |   | 18 | 86 | 141 | 136 | 0.189 | -0.134 | 0.119 | -0.088 | -0.054 |
|---|
| PKN1 |   | 5 | 360 | 1674 | 1634 | 0.150 | -0.143 | -0.011 | 0.084 | -0.002 |
|---|
| SERPINB2 |   | 2 | 743 | 764 | 779 | 0.152 | -0.030 | 0.034 | -0.168 | -0.096 |
|---|
| RB1 |   | 31 | 43 | 1 | 22 | 0.013 | -0.051 | 0.057 | 0.100 | -0.082 |
|---|
| SC4MOL |   | 1 | 1195 | 1674 | 1690 | -0.151 | 0.202 | undef | 0.172 | undef |
|---|
| C17orf81 |   | 2 | 743 | 1674 | 1659 | -0.115 | -0.013 | -0.201 | -0.047 | 0.356 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| GPATCH8 |   | 8 | 222 | 614 | 599 | -0.143 | 0.014 | 0.019 | -0.070 | 0.061 |
|---|
| PTPN11 |   | 20 | 81 | 83 | 94 | -0.152 | 0.014 | 0.066 | 0.304 | 0.089 |
|---|
| INSM1 |   | 23 | 66 | 570 | 545 | 0.135 | -0.184 | -0.188 | 0.237 | -0.032 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| IGF2BP1 |   | 15 | 111 | 366 | 359 | 0.135 | -0.036 | undef | 0.190 | undef |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| SKP2 |   | 29 | 45 | 1 | 23 | 0.092 | -0.043 | 0.237 | undef | -0.009 |
|---|
| ADRB2 |   | 1 | 1195 | 1674 | 1690 | -0.001 | -0.008 | 0.045 | 0.013 | 0.037 |
|---|
| NR2F1 |   | 25 | 59 | 83 | 92 | -0.252 | 0.113 | 0.133 | -0.116 | 0.207 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| TCF7L2 |   | 17 | 95 | 366 | 358 | -0.021 | 0.143 | 0.280 | undef | 0.101 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| PKN3 |   | 18 | 86 | 179 | 187 | -0.181 | 0.009 | undef | 0.239 | undef |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| RAB31 |   | 2 | 743 | 1650 | 1635 | 0.028 | 0.073 | 0.067 | 0.018 | 0.338 |
|---|
| VWF |   | 5 | 360 | 525 | 518 | -0.091 | 0.072 | 0.152 | 0.127 | 0.100 |
|---|
GO Enrichment output for subnetwork 6558 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| carbohydrate homeostasis | GO:0033500 |  | 2.82E-11 | 6.889E-08 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 7.441E-11 | 9.09E-08 |
|---|
| organ growth | GO:0035265 |  | 8.171E-11 | 6.654E-08 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 1.501E-10 | 9.165E-08 |
|---|
| positive regulation of hormone secretion | GO:0046887 |  | 5.178E-10 | 2.53E-07 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 6.142E-10 | 2.501E-07 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 1.137E-09 | 3.969E-07 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.767E-09 | 5.396E-07 |
|---|
| activation of MAPK activity | GO:0000187 |  | 2.635E-09 | 7.153E-07 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.805E-09 | 6.853E-07 |
|---|
| regulation of insulin secretion | GO:0050796 |  | 3.292E-09 | 7.312E-07 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| carbohydrate homeostasis | GO:0033500 |  | 3.526E-11 | 8.483E-08 |
|---|
| organ growth | GO:0035265 |  | 1.091E-10 | 1.313E-07 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.281E-10 | 1.027E-07 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 1.891E-10 | 1.137E-07 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 5.842E-10 | 2.811E-07 |
|---|
| positive regulation of hormone secretion | GO:0046887 |  | 7.293E-10 | 2.925E-07 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 1.125E-09 | 3.868E-07 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.292E-09 | 3.884E-07 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.461E-09 | 9.253E-07 |
|---|
| activation of MAPK activity | GO:0000187 |  | 4.096E-09 | 9.856E-07 |
|---|
| interphase | GO:0051325 |  | 4.569E-09 | 9.994E-07 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.621E-10 | 3.728E-07 |
|---|
| organ growth | GO:0035265 |  | 2.05E-10 | 2.358E-07 |
|---|
| carbohydrate homeostasis | GO:0033500 |  | 1.365E-09 | 1.046E-06 |
|---|
| activation of MAPK activity | GO:0000187 |  | 3.724E-09 | 2.141E-06 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 7.239E-09 | 3.33E-06 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 7.943E-09 | 3.045E-06 |
|---|
| negative regulation of transport | GO:0051051 |  | 2.096E-08 | 6.888E-06 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 2.338E-08 | 6.721E-06 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 2.371E-08 | 6.059E-06 |
|---|
| regulation of multicellular organism growth | GO:0040014 |  | 2.917E-08 | 6.71E-06 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 3.904E-08 | 8.162E-06 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell cycle checkpoint | GO:0000075 |  | 2.745E-07 | 5.06E-04 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 8.195E-07 | 7.551E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.045E-06 | 6.417E-04 |
|---|
| positive regulation of protein kinase activity | GO:0045860 |  | 1.563E-06 | 7.202E-04 |
|---|
| positive regulation of transferase activity | GO:0051347 |  | 1.828E-06 | 6.738E-04 |
|---|
| negative regulation of cell growth | GO:0030308 |  | 2.098E-06 | 6.445E-04 |
|---|
| negative regulation of cell size | GO:0045792 |  | 2.46E-06 | 6.478E-04 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 2.87E-06 | 6.611E-04 |
|---|
| T cell lineage commitment | GO:0002360 |  | 2.96E-06 | 6.062E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 3.849E-06 | 7.093E-04 |
|---|
| response to hormone stimulus | GO:0009725 |  | 4.334E-06 | 7.262E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.621E-10 | 3.728E-07 |
|---|
| organ growth | GO:0035265 |  | 2.05E-10 | 2.358E-07 |
|---|
| carbohydrate homeostasis | GO:0033500 |  | 1.365E-09 | 1.046E-06 |
|---|
| activation of MAPK activity | GO:0000187 |  | 3.724E-09 | 2.141E-06 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 7.239E-09 | 3.33E-06 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 7.943E-09 | 3.045E-06 |
|---|
| negative regulation of transport | GO:0051051 |  | 2.096E-08 | 6.888E-06 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 2.338E-08 | 6.721E-06 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 2.371E-08 | 6.059E-06 |
|---|
| regulation of multicellular organism growth | GO:0040014 |  | 2.917E-08 | 6.71E-06 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 3.904E-08 | 8.162E-06 |
|---|



