Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6434
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7559 | 3.389e-03 | 5.460e-04 | 7.654e-01 | 1.416e-06 |
|---|
| Loi | 0.2298 | 8.107e-02 | 1.157e-02 | 4.302e-01 | 4.036e-04 |
|---|
| Schmidt | 0.6726 | 0.000e+00 | 0.000e+00 | 4.578e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7905 | 0.000e+00 | 0.000e+00 | 3.497e-03 | 0.000e+00 |
|---|
| Wang | 0.2607 | 2.794e-03 | 4.487e-02 | 4.012e-01 | 5.029e-05 |
|---|
Expression data for subnetwork 6434 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
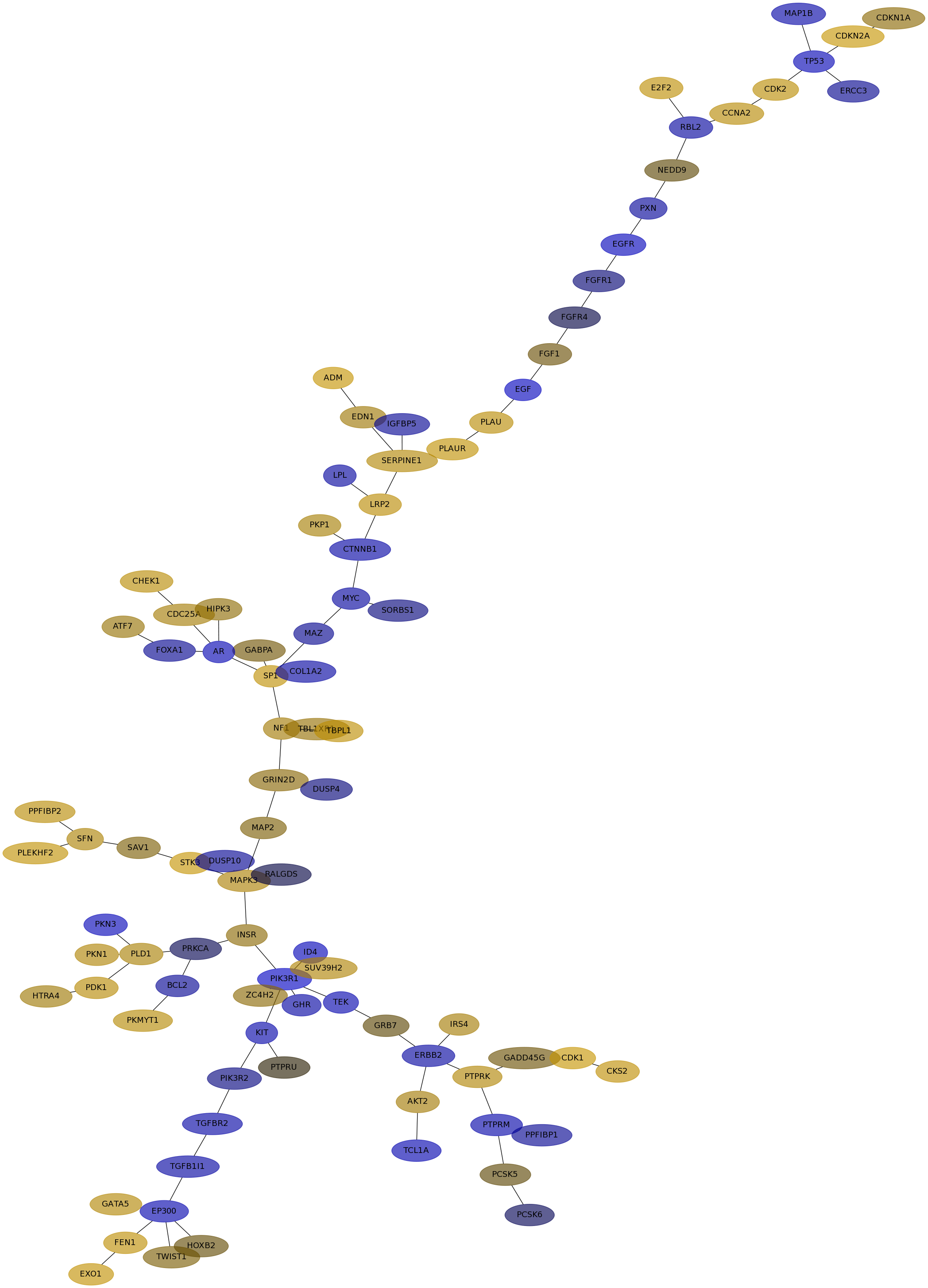
Score for each gene in subnetwork 6434 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| TEK |   | 18 | 86 | 83 | 95 | -0.149 | -0.049 | 0.174 | 0.015 | 0.113 |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| NF1 |   | 9 | 196 | 552 | 541 | 0.110 | -0.052 | -0.032 | 0.090 | -0.094 |
|---|
| ERCC3 |   | 2 | 743 | 478 | 490 | -0.070 | 0.018 | -0.075 | -0.212 | 0.152 |
|---|
| EGF |   | 12 | 140 | 638 | 620 | -0.212 | 0.182 | -0.276 | -0.060 | 0.005 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| PPFIBP2 |   | 2 | 743 | 2024 | 2002 | 0.188 | 0.320 | -0.112 | 0.053 | 0.013 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| ID4 |   | 35 | 34 | 236 | 223 | -0.179 | 0.137 | 0.241 | undef | 0.036 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| CCNA2 |   | 21 | 73 | 179 | 183 | 0.176 | -0.053 | 0.188 | -0.078 | 0.246 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| PLAU |   | 12 | 140 | 179 | 192 | 0.187 | 0.153 | 0.141 | 0.072 | 0.192 |
|---|
| PPFIBP1 |   | 2 | 743 | 1047 | 1055 | -0.073 | 0.207 | -0.072 | 0.085 | -0.016 |
|---|
| PIK3R2 |   | 4 | 440 | 141 | 165 | -0.047 | 0.015 | -0.147 | undef | -0.135 |
|---|
| DUSP10 |   | 1 | 1195 | 2024 | 2033 | -0.076 | 0.158 | -0.206 | undef | 0.007 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| FEN1 |   | 2 | 743 | 2024 | 2002 | 0.210 | -0.132 | 0.008 | 0.002 | 0.028 |
|---|
| PTPRK |   | 14 | 117 | 83 | 96 | 0.156 | 0.141 | -0.094 | 0.048 | -0.024 |
|---|
| CKS2 |   | 9 | 196 | 928 | 903 | 0.214 | -0.220 | 0.089 | -0.003 | -0.056 |
|---|
| PDK1 |   | 10 | 167 | 1 | 40 | 0.183 | -0.138 | -0.041 | -0.265 | -0.126 |
|---|
| GABPA |   | 23 | 66 | 696 | 665 | 0.033 | -0.183 | 0.066 | 0.141 | -0.161 |
|---|
| PTPRU |   | 1 | 1195 | 2024 | 2033 | 0.005 | 0.180 | 0.171 | undef | -0.085 |
|---|
| PTPRM |   | 1 | 1195 | 2024 | 2033 | -0.132 | 0.049 | 0.101 | undef | 0.066 |
|---|
| AKT2 |   | 28 | 47 | 412 | 394 | 0.110 | -0.118 | -0.003 | 0.088 | -0.092 |
|---|
| PKMYT1 |   | 7 | 256 | 1510 | 1463 | 0.175 | -0.143 | 0.046 | 0.105 | -0.070 |
|---|
| DUSP4 |   | 1 | 1195 | 2024 | 2033 | -0.040 | 0.173 | -0.035 | 0.113 | -0.207 |
|---|
| SAV1 |   | 1 | 1195 | 2024 | 2033 | 0.044 | 0.088 | 0.037 | undef | -0.091 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| FGFR1 |   | 26 | 52 | 638 | 615 | -0.036 | 0.200 | 0.087 | undef | -0.015 |
|---|
| PLEKHF2 |   | 1 | 1195 | 2024 | 2033 | 0.232 | -0.095 | -0.032 | undef | 0.016 |
|---|
| ADM |   | 10 | 167 | 682 | 653 | 0.255 | 0.187 | 0.091 | 0.169 | 0.212 |
|---|
| EXO1 |   | 1 | 1195 | 2024 | 2033 | 0.236 | -0.193 | -0.008 | 0.040 | -0.020 |
|---|
| FGF1 |   | 4 | 440 | 1604 | 1572 | 0.027 | undef | undef | 0.090 | undef |
|---|
| MAP2 |   | 3 | 557 | 552 | 558 | 0.044 | 0.045 | 0.014 | undef | -0.057 |
|---|
| HTRA4 |   | 8 | 222 | 83 | 101 | 0.102 | undef | undef | undef | undef |
|---|
| TCL1A |   | 3 | 557 | 1450 | 1430 | -0.152 | -0.166 | 0.090 | undef | -0.066 |
|---|
| SERPINE1 |   | 8 | 222 | 179 | 200 | 0.160 | 0.200 | 0.056 | 0.070 | 0.030 |
|---|
| LPL |   | 13 | 124 | 236 | 230 | -0.102 | -0.062 | 0.292 | 0.087 | -0.105 |
|---|
| GATA5 |   | 8 | 222 | 366 | 373 | 0.161 | -0.031 | undef | undef | undef |
|---|
| E2F2 |   | 13 | 124 | 1 | 35 | 0.212 | -0.047 | 0.162 | -0.131 | -0.092 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| PCSK5 |   | 2 | 743 | 727 | 734 | 0.021 | 0.074 | 0.121 | undef | 0.092 |
|---|
| NEDD9 |   | 1 | 1195 | 2024 | 2033 | 0.020 | 0.023 | 0.032 | -0.046 | 0.222 |
|---|
| ATF7 |   | 28 | 47 | 179 | 175 | 0.085 | 0.115 | -0.088 | 0.172 | -0.092 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| LRP2 |   | 3 | 557 | 1102 | 1098 | 0.193 | -0.042 | 0.001 | -0.138 | 0.125 |
|---|
| RALGDS |   | 5 | 360 | 2024 | 1972 | -0.011 | 0.199 | 0.033 | undef | -0.009 |
|---|
| CHEK1 |   | 18 | 86 | 141 | 136 | 0.189 | -0.134 | 0.119 | -0.088 | -0.054 |
|---|
| PKN1 |   | 5 | 360 | 1674 | 1634 | 0.150 | -0.143 | -0.011 | 0.084 | -0.002 |
|---|
| TGFBR2 |   | 7 | 256 | 525 | 515 | -0.115 | -0.028 | 0.136 | 0.105 | -0.190 |
|---|
| RBL2 |   | 1 | 1195 | 2024 | 2033 | -0.101 | 0.012 | -0.100 | -0.050 | 0.167 |
|---|
| HOXB2 |   | 33 | 37 | 318 | 289 | 0.024 | -0.143 | 0.076 | 0.247 | -0.184 |
|---|
| SUV39H2 |   | 8 | 222 | 318 | 325 | 0.150 | -0.097 | -0.057 | undef | 0.028 |
|---|
| STK3 |   | 4 | 440 | 141 | 165 | 0.254 | 0.007 | 0.106 | 0.326 | 0.198 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| MAZ |   | 2 | 743 | 318 | 346 | -0.068 | -0.208 | -0.157 | undef | -0.175 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| PCSK6 |   | 1 | 1195 | 2024 | 2033 | -0.017 | 0.107 | 0.217 | -0.176 | -0.035 |
|---|
| FGFR4 |   | 21 | 73 | 179 | 183 | -0.011 | 0.188 | -0.089 | -0.018 | -0.049 |
|---|
| CDC25A |   | 8 | 222 | 570 | 555 | 0.113 | -0.170 | 0.151 | -0.301 | -0.003 |
|---|
| PXN |   | 12 | 140 | 1 | 36 | -0.088 | 0.284 | -0.022 | 0.060 | 0.114 |
|---|
| TBL1XR1 |   | 3 | 557 | 552 | 558 | 0.084 | -0.064 | undef | undef | undef |
|---|
| PLAUR |   | 5 | 360 | 696 | 680 | 0.232 | 0.083 | 0.022 | -0.030 | 0.108 |
|---|
| ZC4H2 |   | 17 | 95 | 570 | 546 | 0.065 | 0.061 | 0.304 | -0.072 | 0.059 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| EDN1 |   | 6 | 301 | 682 | 669 | 0.100 | 0.126 | 0.249 | -0.086 | 0.121 |
|---|
| IRS4 |   | 32 | 40 | 236 | 224 | 0.115 | -0.111 | -0.088 | 0.096 | 0.142 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| MAPK3 |   | 6 | 301 | 141 | 152 | 0.137 | -0.102 | -0.126 | undef | -0.086 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| PKN3 |   | 18 | 86 | 179 | 187 | -0.181 | 0.009 | undef | 0.239 | undef |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| TBPL1 |   | 3 | 557 | 236 | 265 | 0.211 | -0.037 | 0.023 | 0.061 | 0.217 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| GRIN2D |   | 13 | 124 | 552 | 522 | 0.058 | 0.155 | 0.000 | undef | undef |
|---|
| PLD1 |   | 17 | 95 | 1 | 29 | 0.132 | 0.033 | -0.158 | -0.062 | 0.123 |
|---|
| PKP1 |   | 1 | 1195 | 2024 | 2033 | 0.123 | 0.104 | 0.127 | -0.115 | 0.011 |
|---|
| COL1A2 |   | 7 | 256 | 552 | 542 | -0.110 | 0.207 | 0.046 | 0.235 | 0.097 |
|---|
| CDK2 |   | 31 | 43 | 179 | 174 | 0.196 | -0.096 | 0.109 | 0.001 | 0.362 |
|---|
| HIPK3 |   | 9 | 196 | 1461 | 1417 | 0.069 | 0.005 | -0.027 | undef | 0.175 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
| SFN |   | 25 | 59 | 478 | 454 | 0.141 | 0.270 | 0.023 | -0.102 | 0.101 |
|---|
| FOXA1 |   | 27 | 50 | 179 | 176 | -0.072 | 0.071 | -0.015 | 0.214 | 0.005 |
|---|
GO Enrichment output for subnetwork 6434 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.795E-12 | 1.416E-08 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 7.731E-10 | 9.443E-07 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.076E-09 | 8.762E-07 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.227E-09 | 7.497E-07 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 2.037E-09 | 9.955E-07 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 3.35E-09 | 1.364E-06 |
|---|
| nerve growth factor processing | GO:0032455 |  | 1.043E-08 | 3.638E-06 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.585E-08 | 4.84E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.264E-08 | 6.144E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.518E-08 | 6.151E-06 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.673E-08 | 5.937E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.128E-11 | 2.713E-08 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.093E-09 | 1.315E-06 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.489E-09 | 1.194E-06 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.705E-09 | 1.026E-06 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 2.871E-09 | 1.382E-06 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 5.05E-09 | 2.025E-06 |
|---|
| nerve growth factor processing | GO:0032455 |  | 1.378E-08 | 4.735E-06 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 2.842E-08 | 8.548E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.428E-08 | 9.165E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.616E-08 | 8.7E-06 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 3.771E-08 | 8.249E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.038E-10 | 1.619E-06 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.987E-09 | 2.286E-06 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.334E-09 | 1.79E-06 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 3.101E-09 | 1.783E-06 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 3.715E-09 | 1.709E-06 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 8.409E-09 | 3.223E-06 |
|---|
| nerve growth factor processing | GO:0032455 |  | 3.189E-08 | 1.048E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.257E-08 | 1.799E-05 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 7.333E-08 | 1.874E-05 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.061E-07 | 2.44E-05 |
|---|
| response to UV | GO:0009411 |  | 1.716E-07 | 3.589E-05 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of MAP kinase activity | GO:0043405 |  | 2.082E-09 | 3.836E-06 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 4.001E-09 | 3.687E-06 |
|---|
| positive regulation of protein kinase activity | GO:0045860 |  | 2.284E-08 | 1.403E-05 |
|---|
| positive regulation of transferase activity | GO:0051347 |  | 2.803E-08 | 1.292E-05 |
|---|
| response to radiation | GO:0009314 |  | 5.824E-08 | 2.147E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 5.835E-08 | 1.792E-05 |
|---|
| regulation of mitotic cell cycle | GO:0007346 |  | 6.547E-08 | 1.724E-05 |
|---|
| regulation of cell migration | GO:0030334 |  | 8.227E-08 | 1.895E-05 |
|---|
| response to UV | GO:0009411 |  | 9.088E-08 | 1.861E-05 |
|---|
| response to light stimulus | GO:0009416 |  | 1.622E-07 | 2.989E-05 |
|---|
| regulation of locomotion | GO:0040012 |  | 1.919E-07 | 3.215E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.038E-10 | 1.619E-06 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.987E-09 | 2.286E-06 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.334E-09 | 1.79E-06 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 3.101E-09 | 1.783E-06 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 3.715E-09 | 1.709E-06 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 8.409E-09 | 3.223E-06 |
|---|
| nerve growth factor processing | GO:0032455 |  | 3.189E-08 | 1.048E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.257E-08 | 1.799E-05 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 7.333E-08 | 1.874E-05 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.061E-07 | 2.44E-05 |
|---|
| response to UV | GO:0009411 |  | 1.716E-07 | 3.589E-05 |
|---|



