Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6390
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7565 | 3.350e-03 | 5.380e-04 | 7.636e-01 | 1.376e-06 |
|---|
| Loi | 0.2295 | 8.159e-02 | 1.171e-02 | 4.314e-01 | 4.122e-04 |
|---|
| Schmidt | 0.6724 | 0.000e+00 | 0.000e+00 | 4.602e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7906 | 0.000e+00 | 0.000e+00 | 3.494e-03 | 0.000e+00 |
|---|
| Wang | 0.2604 | 2.838e-03 | 4.526e-02 | 4.030e-01 | 5.177e-05 |
|---|
Expression data for subnetwork 6390 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
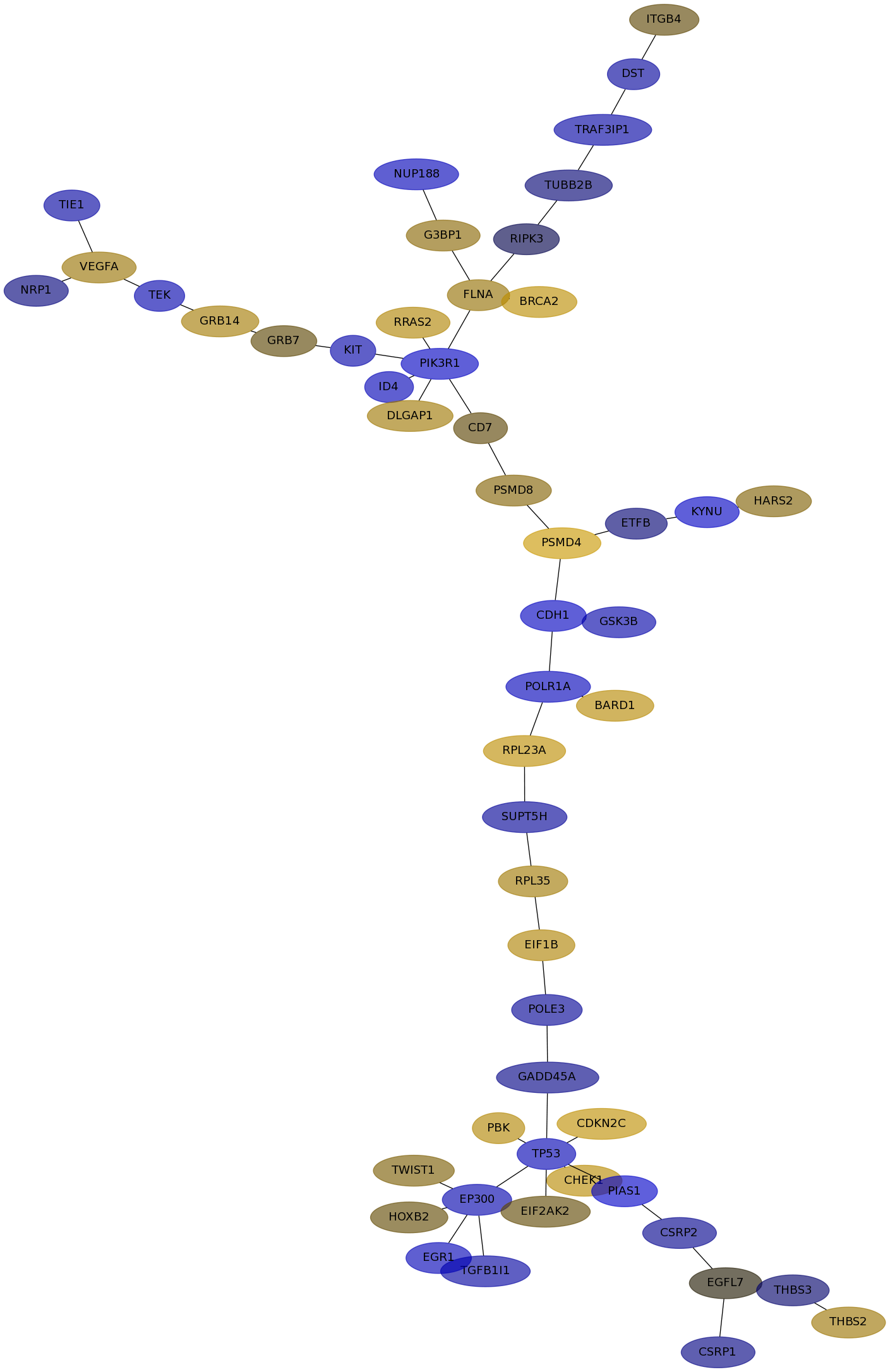
Score for each gene in subnetwork 6390 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| TEK |   | 18 | 86 | 83 | 95 | -0.149 | -0.049 | 0.174 | 0.015 | 0.113 |
|---|
| NUP188 |   | 8 | 222 | 1116 | 1079 | -0.187 | 0.094 | 0.058 | 0.161 | -0.070 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| GRB14 |   | 24 | 62 | 179 | 181 | 0.113 | -0.129 | 0.143 | -0.098 | 0.138 |
|---|
| POLR1A |   | 6 | 301 | 457 | 452 | -0.193 | -0.066 | undef | 0.093 | undef |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| CHEK1 |   | 18 | 86 | 141 | 136 | 0.189 | -0.134 | 0.119 | -0.088 | -0.054 |
|---|
| ID4 |   | 35 | 34 | 236 | 223 | -0.179 | 0.137 | 0.241 | undef | 0.036 |
|---|
| THBS3 |   | 1 | 1195 | 2207 | 2211 | -0.030 | 0.086 | 0.270 | 0.186 | -0.005 |
|---|
| CD7 |   | 2 | 743 | 1102 | 1102 | 0.020 | -0.151 | -0.165 | 0.027 | -0.073 |
|---|
| EIF1B |   | 8 | 222 | 1576 | 1529 | 0.149 | 0.075 | 0.332 | -0.017 | -0.070 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| HOXB2 |   | 33 | 37 | 318 | 289 | 0.024 | -0.143 | 0.076 | 0.247 | -0.184 |
|---|
| ETFB |   | 2 | 743 | 2003 | 1980 | -0.037 | 0.058 | 0.081 | -0.105 | -0.065 |
|---|
| RPL23A |   | 5 | 360 | 842 | 828 | 0.208 | -0.007 | 0.024 | -0.242 | 0.055 |
|---|
| DST |   | 22 | 69 | 1 | 25 | -0.094 | 0.343 | 0.065 | -0.062 | 0.194 |
|---|
| CDKN2C |   | 16 | 104 | 236 | 227 | 0.216 | -0.106 | 0.202 | -0.024 | -0.081 |
|---|
| PIAS1 |   | 2 | 743 | 2090 | 2082 | -0.270 | 0.096 | 0.157 | -0.213 | -0.083 |
|---|
| EIF2AK2 |   | 9 | 196 | 648 | 629 | 0.023 | -0.022 | -0.134 | 0.096 | -0.303 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| HARS2 |   | 3 | 557 | 1268 | 1255 | 0.049 | 0.360 | -0.168 | undef | -0.199 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| POLE3 |   | 17 | 95 | 141 | 138 | -0.083 | -0.033 | 0.231 | 0.096 | 0.011 |
|---|
| RRAS2 |   | 7 | 256 | 1681 | 1633 | 0.155 | 0.076 | 0.289 | 0.044 | 0.031 |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| DLGAP1 |   | 2 | 743 | 2207 | 2182 | 0.103 | 0.078 | 0.102 | -0.072 | -0.159 |
|---|
| RPL35 |   | 8 | 222 | 1323 | 1280 | 0.108 | -0.012 | 0.370 | -0.145 | -0.220 |
|---|
| CSRP1 |   | 2 | 743 | 1975 | 1953 | -0.053 | 0.224 | 0.088 | 0.049 | 0.080 |
|---|
| THBS2 |   | 3 | 557 | 1903 | 1868 | 0.094 | 0.221 | 0.060 | 0.241 | 0.154 |
|---|
| PBK |   | 2 | 743 | 2207 | 2182 | 0.163 | -0.164 | 0.107 | -0.067 | -0.095 |
|---|
| RIPK3 |   | 19 | 85 | 179 | 186 | -0.014 | 0.040 | undef | 0.173 | undef |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| PSMD4 |   | 11 | 148 | 296 | 282 | 0.285 | -0.243 | -0.043 | 0.087 | -0.045 |
|---|
| SUPT5H |   | 6 | 301 | 1323 | 1281 | -0.085 | 0.007 | -0.298 | 0.002 | -0.068 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| GADD45A |   | 11 | 148 | 1351 | 1308 | -0.057 | 0.076 | -0.099 | 0.109 | 0.093 |
|---|
| PSMD8 |   | 1 | 1195 | 2207 | 2211 | 0.052 | -0.027 | -0.080 | 0.123 | 0.027 |
|---|
| KYNU |   | 1 | 1195 | 2207 | 2211 | -0.245 | 0.090 | -0.294 | 0.107 | 0.047 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| TRAF3IP1 |   | 9 | 196 | 83 | 99 | -0.113 | -0.063 | -0.088 | 0.276 | undef |
|---|
| TUBB2B |   | 13 | 124 | 141 | 141 | -0.036 | 0.304 | 0.045 | 0.187 | 0.139 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| TIE1 |   | 18 | 86 | 366 | 357 | -0.108 | 0.005 | 0.141 | -0.108 | 0.252 |
|---|
| EGFL7 |   | 1 | 1195 | 2207 | 2211 | 0.004 | 0.087 | 0.128 | undef | 0.175 |
|---|
| ITGB4 |   | 35 | 34 | 1 | 20 | 0.021 | 0.313 | -0.134 | 0.080 | -0.114 |
|---|
| NRP1 |   | 8 | 222 | 1222 | 1186 | -0.043 | 0.025 | 0.201 | 0.029 | 0.071 |
|---|
| G3BP1 |   | 7 | 256 | 1116 | 1082 | 0.060 | -0.077 | -0.017 | 0.155 | -0.004 |
|---|
| CSRP2 |   | 1 | 1195 | 2207 | 2211 | -0.067 | -0.015 | 0.182 | 0.194 | 0.046 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
GO Enrichment output for subnetwork 6390 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 2.736E-07 | 6.685E-04 |
|---|
| regulation of granulocyte macrophage colony-stimulating factor production | GO:0032645 |  | 3.459E-07 | 4.226E-04 |
|---|
| T cell activation | GO:0042110 |  | 3.685E-07 | 3.001E-04 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 4.363E-07 | 2.665E-04 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 5.186E-07 | 2.534E-04 |
|---|
| response to vitamin | GO:0033273 |  | 5.186E-07 | 2.112E-04 |
|---|
| positive regulation of T-helper 1 cell differentiation | GO:0045627 |  | 6.902E-07 | 2.409E-04 |
|---|
| reciprocal meiotic recombination | GO:0007131 |  | 7.791E-07 | 2.379E-04 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 7.791E-07 | 2.115E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 9.193E-07 | 2.246E-04 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.205E-06 | 2.676E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 3.592E-07 | 8.642E-04 |
|---|
| T cell activation | GO:0042110 |  | 4.081E-07 | 4.91E-04 |
|---|
| regulation of granulocyte macrophage colony-stimulating factor production | GO:0032645 |  | 4.097E-07 | 3.286E-04 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 5.724E-07 | 3.443E-04 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 6.473E-07 | 3.115E-04 |
|---|
| response to vitamin | GO:0033273 |  | 6.473E-07 | 2.596E-04 |
|---|
| reciprocal meiotic recombination | GO:0007131 |  | 7.975E-07 | 2.741E-04 |
|---|
| positive regulation of T-helper 1 cell differentiation | GO:0045627 |  | 8.173E-07 | 2.458E-04 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 9.721E-07 | 2.599E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.051E-06 | 2.528E-04 |
|---|
| interphase | GO:0051325 |  | 1.262E-06 | 2.76E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 9.289E-07 | 2.136E-03 |
|---|
| regulation of granulocyte macrophage colony-stimulating factor production | GO:0032645 |  | 1.31E-06 | 1.506E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 1.485E-06 | 1.139E-03 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 2.366E-06 | 1.36E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 2.4E-06 | 1.104E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 2.61E-06 | 1E-03 |
|---|
| ER overload response | GO:0006983 |  | 2.61E-06 | 8.574E-04 |
|---|
| positive regulation of T-helper 1 cell differentiation | GO:0045627 |  | 2.61E-06 | 7.503E-04 |
|---|
| response to vitamin | GO:0033273 |  | 2.988E-06 | 7.635E-04 |
|---|
| T cell activation | GO:0042110 |  | 3.233E-06 | 7.437E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 3.676E-06 | 7.686E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 3.635E-07 | 6.7E-04 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 4.229E-07 | 3.897E-04 |
|---|
| cell cycle arrest | GO:0007050 |  | 4.754E-07 | 2.92E-04 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 5.473E-07 | 2.522E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 9.081E-07 | 3.347E-04 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 1.131E-06 | 3.474E-04 |
|---|
| response to vitamin | GO:0033273 |  | 1.718E-06 | 4.523E-04 |
|---|
| ER-nuclear signaling pathway | GO:0006984 |  | 1.718E-06 | 3.958E-04 |
|---|
| positive regulation of transcription factor import into nucleus | GO:0042993 |  | 4.49E-06 | 9.195E-04 |
|---|
| ER overload response | GO:0006983 |  | 4.49E-06 | 8.275E-04 |
|---|
| T cell activation | GO:0042110 |  | 1.292E-05 | 2.164E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 9.289E-07 | 2.136E-03 |
|---|
| regulation of granulocyte macrophage colony-stimulating factor production | GO:0032645 |  | 1.31E-06 | 1.506E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 1.485E-06 | 1.139E-03 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 2.366E-06 | 1.36E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 2.4E-06 | 1.104E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 2.61E-06 | 1E-03 |
|---|
| ER overload response | GO:0006983 |  | 2.61E-06 | 8.574E-04 |
|---|
| positive regulation of T-helper 1 cell differentiation | GO:0045627 |  | 2.61E-06 | 7.503E-04 |
|---|
| response to vitamin | GO:0033273 |  | 2.988E-06 | 7.635E-04 |
|---|
| T cell activation | GO:0042110 |  | 3.233E-06 | 7.437E-04 |
|---|
| response to endoplasmic reticulum stress | GO:0034976 |  | 3.676E-06 | 7.686E-04 |
|---|



