Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6388
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7568 | 3.333e-03 | 5.340e-04 | 7.629e-01 | 1.358e-06 |
|---|
| Loi | 0.2294 | 8.176e-02 | 1.175e-02 | 4.318e-01 | 4.150e-04 |
|---|
| Schmidt | 0.6724 | 0.000e+00 | 0.000e+00 | 4.601e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7906 | 0.000e+00 | 0.000e+00 | 3.496e-03 | 0.000e+00 |
|---|
| Wang | 0.2604 | 2.835e-03 | 4.523e-02 | 4.029e-01 | 5.166e-05 |
|---|
Expression data for subnetwork 6388 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
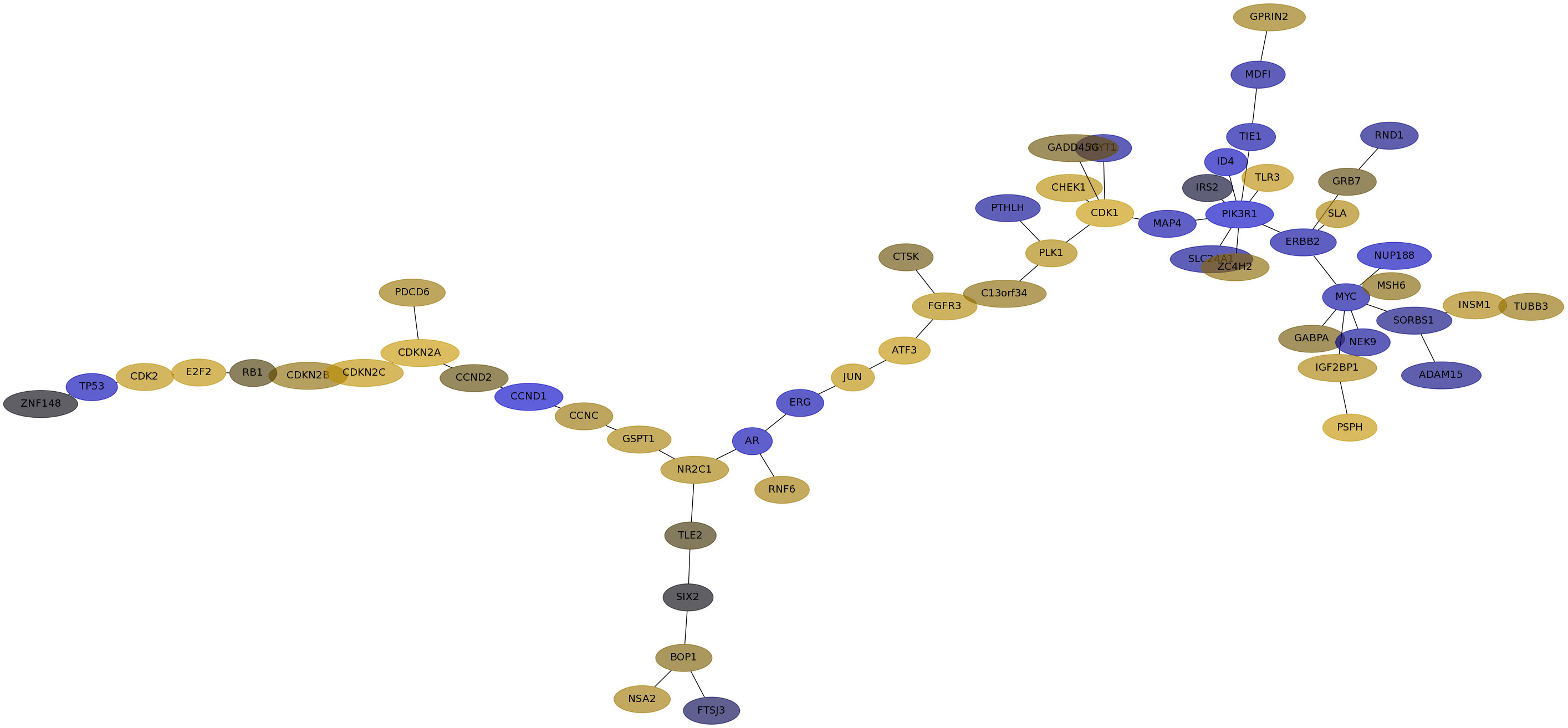
Score for each gene in subnetwork 6388 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| CDKN2B |   | 7 | 256 | 236 | 237 | 0.066 | 0.050 | -0.028 | 0.039 | 0.153 |
|---|
| CCNC |   | 5 | 360 | 570 | 562 | 0.085 | -0.053 | undef | 0.028 | undef |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| NUP188 |   | 8 | 222 | 1116 | 1079 | -0.187 | 0.094 | 0.058 | 0.161 | -0.070 |
|---|
| PTHLH |   | 2 | 743 | 478 | 490 | -0.070 | 0.111 | 0.047 | -0.033 | -0.091 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CHEK1 |   | 18 | 86 | 141 | 136 | 0.189 | -0.134 | 0.119 | -0.088 | -0.054 |
|---|
| ID4 |   | 35 | 34 | 236 | 223 | -0.179 | 0.137 | 0.241 | undef | 0.036 |
|---|
| SLA |   | 2 | 743 | 1938 | 1922 | 0.138 | -0.195 | -0.058 | undef | -0.012 |
|---|
| RND1 |   | 6 | 301 | 236 | 239 | -0.046 | 0.085 | 0.022 | 0.082 | 0.113 |
|---|
| JUN |   | 26 | 52 | 179 | 177 | 0.200 | 0.051 | 0.220 | undef | 0.200 |
|---|
| CCND1 |   | 26 | 52 | 1 | 24 | -0.258 | 0.074 | 0.117 | undef | 0.003 |
|---|
| CDKN2C |   | 16 | 104 | 236 | 227 | 0.216 | -0.106 | 0.202 | -0.024 | -0.081 |
|---|
| FGFR3 |   | 9 | 196 | 179 | 195 | 0.160 | 0.071 | 0.018 | -0.050 | -0.111 |
|---|
| RB1 |   | 31 | 43 | 1 | 22 | 0.013 | -0.051 | 0.057 | 0.100 | -0.082 |
|---|
| TLR3 |   | 1 | 1195 | 2040 | 2044 | 0.196 | -0.111 | -0.089 | -0.063 | -0.103 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| GPRIN2 |   | 16 | 104 | 179 | 190 | 0.084 | 0.078 | 0.012 | 0.054 | 0.114 |
|---|
| BOP1 |   | 1 | 1195 | 2040 | 2044 | 0.045 | -0.127 | 0.088 | undef | -0.126 |
|---|
| ADAM15 |   | 3 | 557 | 1803 | 1777 | -0.048 | -0.078 | -0.208 | 0.172 | -0.048 |
|---|
| INSM1 |   | 23 | 66 | 570 | 545 | 0.135 | -0.184 | -0.188 | 0.237 | -0.032 |
|---|
| ZNF148 |   | 1 | 1195 | 2040 | 2044 | -0.001 | 0.161 | 0.077 | undef | 0.161 |
|---|
| IGF2BP1 |   | 15 | 111 | 366 | 359 | 0.135 | -0.036 | undef | 0.190 | undef |
|---|
| CCND2 |   | 5 | 360 | 1492 | 1453 | 0.022 | -0.011 | -0.072 | -0.153 | -0.029 |
|---|
| MYT1 |   | 1 | 1195 | 2040 | 2044 | -0.070 | -0.100 | 0.206 | undef | -0.039 |
|---|
| NR2C1 |   | 8 | 222 | 727 | 703 | 0.121 | 0.039 | 0.119 | -0.197 | -0.026 |
|---|
| FTSJ3 |   | 1 | 1195 | 2040 | 2044 | -0.016 | 0.102 | 0.015 | 0.027 | 0.019 |
|---|
| MSH6 |   | 12 | 140 | 1 | 36 | 0.052 | -0.057 | 0.051 | 0.126 | 0.064 |
|---|
| TLE2 |   | 4 | 440 | 806 | 790 | 0.010 | 0.052 | 0.202 | undef | 0.093 |
|---|
| TUBB3 |   | 7 | 256 | 764 | 756 | 0.078 | 0.130 | 0.073 | 0.207 | -0.079 |
|---|
| MDFI |   | 17 | 95 | 236 | 226 | -0.075 | -0.001 | 0.177 | 0.169 | -0.112 |
|---|
| PLK1 |   | 11 | 148 | 236 | 232 | 0.142 | -0.143 | 0.094 | -0.192 | -0.163 |
|---|
| ERG |   | 9 | 196 | 179 | 195 | -0.134 | -0.026 | 0.235 | -0.027 | 0.182 |
|---|
| GABPA |   | 23 | 66 | 696 | 665 | 0.033 | -0.183 | 0.066 | 0.141 | -0.161 |
|---|
| ZC4H2 |   | 17 | 95 | 570 | 546 | 0.065 | 0.061 | 0.304 | -0.072 | 0.059 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| ATF3 |   | 17 | 95 | 179 | 189 | 0.225 | -0.021 | 0.000 | undef | -0.039 |
|---|
| CTSK |   | 1 | 1195 | 2040 | 2044 | 0.028 | 0.032 | -0.018 | 0.135 | 0.118 |
|---|
| C13orf34 |   | 3 | 557 | 1165 | 1151 | 0.059 | 0.057 | 0.007 | undef | -0.147 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| IRS2 |   | 3 | 557 | 1102 | 1098 | -0.006 | 0.043 | 0.054 | -0.256 | 0.125 |
|---|
| NSA2 |   | 4 | 440 | 806 | 790 | 0.103 | 0.097 | 0.132 | -0.026 | 0.006 |
|---|
| RNF6 |   | 4 | 440 | 590 | 585 | 0.109 | -0.128 | 0.002 | 0.041 | -0.018 |
|---|
| PDCD6 |   | 10 | 167 | 1 | 40 | 0.091 | -0.060 | undef | undef | undef |
|---|
| SLC24A1 |   | 4 | 440 | 1323 | 1295 | -0.068 | 0.109 | 0.178 | -0.020 | 0.056 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| MAP4 |   | 2 | 743 | 1437 | 1433 | -0.115 | 0.217 | 0.221 | 0.086 | 0.268 |
|---|
| NEK9 |   | 2 | 743 | 1659 | 1643 | -0.074 | 0.037 | 0.211 | undef | 0.056 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| GSPT1 |   | 2 | 743 | 1975 | 1953 | 0.123 | -0.024 | 0.036 | 0.037 | -0.040 |
|---|
| TIE1 |   | 18 | 86 | 366 | 357 | -0.108 | 0.005 | 0.141 | -0.108 | 0.252 |
|---|
| CDK2 |   | 31 | 43 | 179 | 174 | 0.196 | -0.096 | 0.109 | 0.001 | 0.362 |
|---|
| E2F2 |   | 13 | 124 | 1 | 35 | 0.212 | -0.047 | 0.162 | -0.131 | -0.092 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
| SIX2 |   | 2 | 743 | 727 | 734 | -0.001 | -0.092 | -0.196 | 0.161 | 0.137 |
|---|
| PSPH |   | 20 | 81 | 366 | 356 | 0.240 | -0.019 | -0.102 | 0.358 | 0.025 |
|---|
GO Enrichment output for subnetwork 6388 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.67E-14 | 4.079E-11 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.346E-11 | 2.865E-08 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.427E-10 | 1.162E-07 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 2.634E-08 | 1.609E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 5.688E-08 | 2.779E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 7.086E-08 | 2.885E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 8.742E-08 | 3.051E-05 |
|---|
| negative regulation of epithelial cell proliferation | GO:0050680 |  | 1.249E-06 | 3.813E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 1.519E-06 | 4.125E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 1.555E-06 | 3.8E-04 |
|---|
| L-serine biosynthetic process | GO:0006564 |  | 1.58E-06 | 3.509E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.478E-14 | 5.961E-11 |
|---|
| interphase | GO:0051325 |  | 3.639E-14 | 4.378E-11 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.005E-11 | 3.212E-08 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.43E-10 | 1.461E-07 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 3.421E-08 | 1.646E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 7.997E-08 | 3.207E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 9.959E-08 | 3.423E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.228E-07 | 3.695E-05 |
|---|
| cell cycle arrest | GO:0007050 |  | 4.503E-07 | 1.204E-04 |
|---|
| L-serine biosynthetic process | GO:0006564 |  | 9.765E-07 | 2.349E-04 |
|---|
| negative regulation of epithelial cell proliferation | GO:0050680 |  | 1.642E-06 | 3.592E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.458E-12 | 1.025E-08 |
|---|
| interphase | GO:0051325 |  | 5.773E-12 | 6.639E-09 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.834E-10 | 1.406E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.448E-08 | 1.408E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.41E-07 | 1.108E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 3.768E-07 | 1.444E-04 |
|---|
| cell cycle arrest | GO:0007050 |  | 9.534E-07 | 3.133E-04 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 2.566E-06 | 7.376E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 3.799E-06 | 9.71E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 3.814E-06 | 8.773E-04 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 4.399E-06 | 9.199E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.686E-10 | 3.108E-07 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.498E-08 | 5.988E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.89E-07 | 3.004E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.818E-07 | 4.523E-04 |
|---|
| negative regulation of cell differentiation | GO:0045596 |  | 1.246E-06 | 4.593E-04 |
|---|
| regulation of cell size | GO:0008361 |  | 1.361E-06 | 4.181E-04 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 2.214E-06 | 5.829E-04 |
|---|
| positive regulation of protein kinase activity | GO:0045860 |  | 5.452E-06 | 1.256E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 5.461E-06 | 1.118E-03 |
|---|
| positive regulation of transferase activity | GO:0051347 |  | 6.236E-06 | 1.149E-03 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 1.528E-05 | 2.559E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.458E-12 | 1.025E-08 |
|---|
| interphase | GO:0051325 |  | 5.773E-12 | 6.639E-09 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.834E-10 | 1.406E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.448E-08 | 1.408E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.41E-07 | 1.108E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 3.768E-07 | 1.444E-04 |
|---|
| cell cycle arrest | GO:0007050 |  | 9.534E-07 | 3.133E-04 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 2.566E-06 | 7.376E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 3.799E-06 | 9.71E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 3.814E-06 | 8.773E-04 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 4.399E-06 | 9.199E-04 |
|---|



