Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6306
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7579 | 3.269e-03 | 5.210e-04 | 7.600e-01 | 1.294e-06 |
|---|
| Loi | 0.2293 | 8.187e-02 | 1.179e-02 | 4.321e-01 | 4.169e-04 |
|---|
| Schmidt | 0.6714 | 0.000e+00 | 0.000e+00 | 4.708e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7906 | 0.000e+00 | 0.000e+00 | 3.487e-03 | 0.000e+00 |
|---|
| Wang | 0.2599 | 2.905e-03 | 4.585e-02 | 4.057e-01 | 5.404e-05 |
|---|
Expression data for subnetwork 6306 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
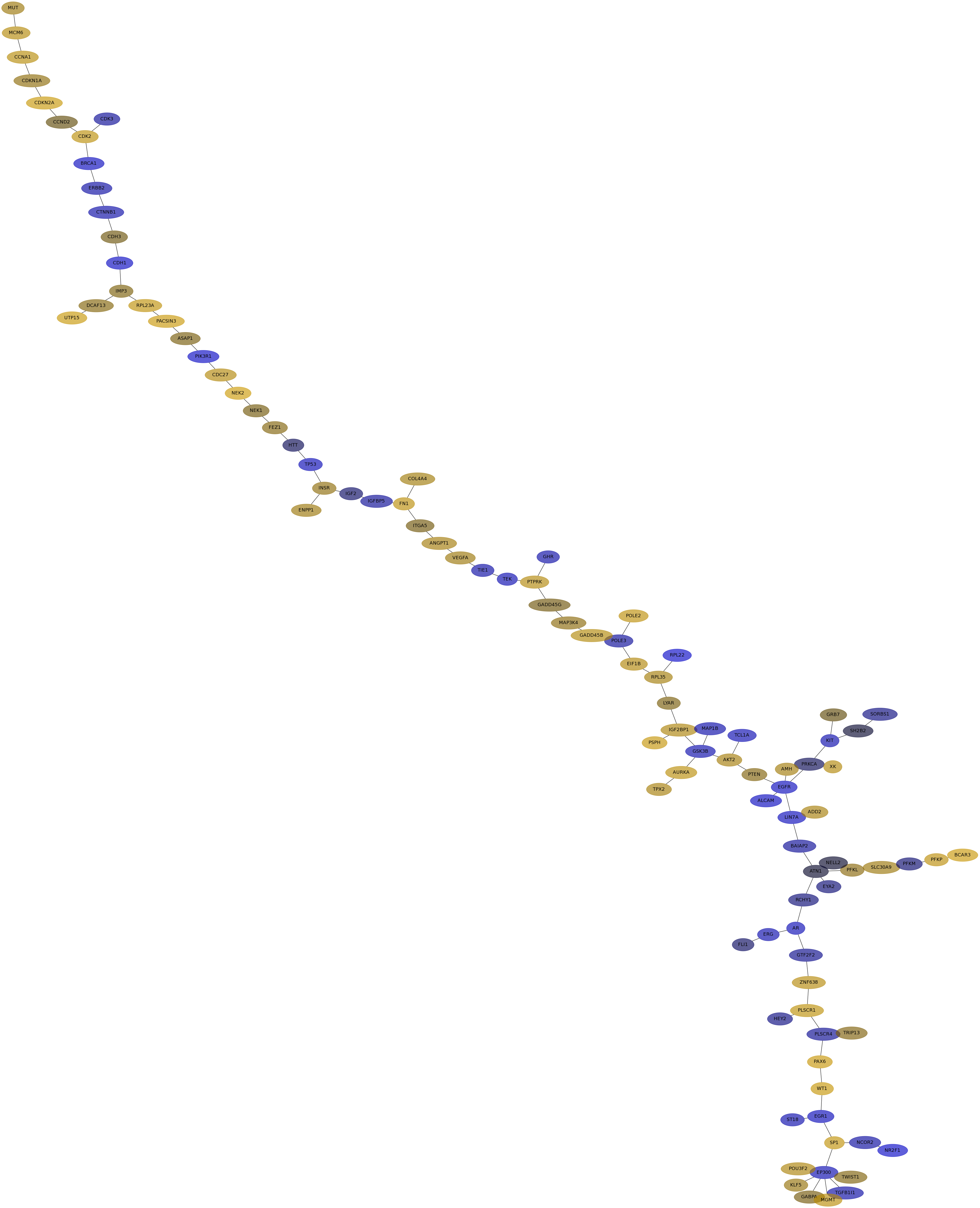
Score for each gene in subnetwork 6306 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| TEK |   | 18 | 86 | 83 | 95 | -0.149 | -0.049 | 0.174 | 0.015 | 0.113 |
|---|
| PAX6 |   | 22 | 69 | 318 | 312 | 0.259 | -0.003 | 0.112 | undef | 0.168 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| TPX2 |   | 5 | 360 | 412 | 416 | 0.114 | -0.121 | -0.018 | undef | -0.056 |
|---|
| RCHY1 |   | 2 | 743 | 2080 | 2055 | -0.035 | 0.007 | 0.245 | undef | -0.026 |
|---|
| ADD2 |   | 8 | 222 | 614 | 599 | 0.113 | -0.022 | -0.061 | 0.270 | 0.180 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| EIF1B |   | 8 | 222 | 1576 | 1529 | 0.149 | 0.075 | 0.332 | -0.017 | -0.070 |
|---|
| CDK3 |   | 7 | 256 | 590 | 570 | -0.076 | 0.189 | -0.041 | -0.089 | 0.082 |
|---|
| RPL23A |   | 5 | 360 | 842 | 828 | 0.208 | -0.007 | 0.024 | -0.242 | 0.055 |
|---|
| HEY2 |   | 8 | 222 | 570 | 555 | -0.045 | -0.021 | 0.398 | -0.024 | -0.058 |
|---|
| NELL2 |   | 7 | 256 | 727 | 714 | -0.005 | 0.138 | 0.421 | -0.027 | 0.074 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| CDH3 |   | 1 | 1195 | 2301 | 2301 | 0.028 | 0.169 | -0.055 | undef | -0.027 |
|---|
| LIN7A |   | 6 | 301 | 366 | 377 | -0.189 | -0.074 | 0.108 | 0.065 | -0.146 |
|---|
| ST18 |   | 16 | 104 | 236 | 227 | -0.131 | 0.028 | -0.088 | 0.186 | 0.151 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| POLE3 |   | 17 | 95 | 141 | 138 | -0.083 | -0.033 | 0.231 | 0.096 | 0.011 |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| TRIP13 |   | 2 | 743 | 1055 | 1059 | 0.043 | -0.199 | -0.017 | undef | -0.146 |
|---|
| PTPRK |   | 14 | 117 | 83 | 96 | 0.156 | 0.141 | -0.094 | 0.048 | -0.024 |
|---|
| ZNF638 |   | 5 | 360 | 806 | 784 | 0.164 | 0.088 | 0.020 | 0.077 | -0.045 |
|---|
| RPL35 |   | 8 | 222 | 1323 | 1280 | 0.108 | -0.012 | 0.370 | -0.145 | -0.220 |
|---|
| GABPA |   | 23 | 66 | 696 | 665 | 0.033 | -0.183 | 0.066 | 0.141 | -0.161 |
|---|
| AKT2 |   | 28 | 47 | 412 | 394 | 0.110 | -0.118 | -0.003 | 0.088 | -0.092 |
|---|
| FEZ1 |   | 8 | 222 | 141 | 149 | 0.047 | -0.020 | 0.154 | 0.165 | -0.128 |
|---|
| BRCA1 |   | 3 | 557 | 1534 | 1516 | -0.201 | -0.078 | -0.087 | -0.166 | 0.168 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| ALCAM |   | 8 | 222 | 1 | 46 | -0.208 | 0.193 | 0.067 | 0.088 | -0.010 |
|---|
| MAP3K4 |   | 1 | 1195 | 2301 | 2301 | 0.059 | -0.015 | 0.000 | -0.169 | -0.092 |
|---|
| ENPP1 |   | 5 | 360 | 1055 | 1034 | 0.088 | 0.129 | -0.026 | undef | 0.039 |
|---|
| GADD45B |   | 8 | 222 | 318 | 325 | 0.158 | 0.122 | 0.067 | 0.060 | 0.212 |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| TCL1A |   | 3 | 557 | 1450 | 1430 | -0.152 | -0.166 | 0.090 | undef | -0.066 |
|---|
| RPL22 |   | 3 | 557 | 1576 | 1548 | -0.266 | 0.105 | 0.059 | -0.143 | 0.112 |
|---|
| ANGPT1 |   | 5 | 360 | 696 | 680 | 0.106 | -0.167 | 0.084 | -0.035 | 0.250 |
|---|
| EYA2 |   | 13 | 124 | 318 | 316 | -0.034 | 0.113 | -0.142 | undef | -0.019 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| PFKP |   | 6 | 301 | 83 | 105 | 0.195 | -0.076 | -0.040 | undef | 0.094 |
|---|
| DCAF13 |   | 2 | 743 | 366 | 395 | 0.049 | -0.211 | undef | -0.060 | undef |
|---|
| PSPH |   | 20 | 81 | 366 | 356 | 0.240 | -0.019 | -0.102 | 0.358 | 0.025 |
|---|
| SLC30A9 |   | 2 | 743 | 83 | 123 | 0.089 | -0.011 | 0.026 | -0.104 | 0.050 |
|---|
| POU3F2 |   | 17 | 95 | 928 | 891 | 0.135 | 0.114 | 0.155 | 0.253 | 0.040 |
|---|
| NCOR2 |   | 32 | 40 | 318 | 290 | -0.096 | 0.211 | -0.163 | 0.158 | 0.114 |
|---|
| MCM6 |   | 1 | 1195 | 2301 | 2301 | 0.162 | -0.181 | 0.021 | 0.010 | 0.199 |
|---|
| PLSCR4 |   | 13 | 124 | 318 | 316 | -0.069 | 0.140 | 0.231 | 0.212 | 0.212 |
|---|
| MUT |   | 1 | 1195 | 2301 | 2301 | 0.088 | 0.056 | -0.051 | 0.294 | 0.183 |
|---|
| FN1 |   | 6 | 301 | 590 | 582 | 0.190 | 0.133 | 0.091 | undef | 0.083 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| LYAR |   | 2 | 743 | 1803 | 1782 | 0.037 | -0.154 | undef | -0.245 | undef |
|---|
| ASAP1 |   | 1 | 1195 | 2301 | 2301 | 0.036 | 0.070 | 0.035 | undef | -0.056 |
|---|
| PFKL |   | 7 | 256 | 83 | 102 | 0.056 | 0.106 | -0.173 | 0.160 | -0.158 |
|---|
| PTEN |   | 27 | 50 | 141 | 135 | 0.045 | 0.055 | 0.017 | 0.183 | -0.041 |
|---|
| AMH |   | 10 | 167 | 366 | 362 | 0.103 | 0.056 | 0.113 | -0.187 | 0.301 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| NEK2 |   | 8 | 222 | 141 | 149 | 0.292 | -0.106 | 0.020 | 0.083 | 0.165 |
|---|
| GTF2F2 |   | 4 | 440 | 957 | 942 | -0.059 | -0.085 | 0.119 | -0.004 | -0.054 |
|---|
| WT1 |   | 33 | 37 | 1 | 21 | 0.254 | -0.039 | 0.036 | undef | 0.172 |
|---|
| FLI1 |   | 2 | 743 | 1 | 66 | -0.021 | 0.018 | 0.067 | -0.175 | 0.009 |
|---|
| IGF2BP1 |   | 15 | 111 | 366 | 359 | 0.135 | -0.036 | undef | 0.190 | undef |
|---|
| CCND2 |   | 5 | 360 | 1492 | 1453 | 0.022 | -0.011 | -0.072 | -0.153 | -0.029 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| ERG |   | 9 | 196 | 179 | 195 | -0.134 | -0.026 | 0.235 | -0.027 | 0.182 |
|---|
| PLSCR1 |   | 10 | 167 | 412 | 408 | 0.202 | -0.159 | 0.091 | undef | -0.105 |
|---|
| COL4A4 |   | 6 | 301 | 83 | 105 | 0.102 | 0.155 | -0.039 | 0.295 | 0.068 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| PACSIN3 |   | 1 | 1195 | 2301 | 2301 | 0.270 | 0.129 | -0.045 | 0.016 | 0.066 |
|---|
| UTP15 |   | 2 | 743 | 366 | 395 | 0.266 | 0.140 | undef | undef | undef |
|---|
| ITGA5 |   | 26 | 52 | 179 | 177 | 0.031 | 0.192 | 0.125 | 0.133 | 0.202 |
|---|
| IMP3 |   | 4 | 440 | 366 | 381 | 0.042 | -0.132 | 0.113 | -0.304 | -0.196 |
|---|
| PFKM |   | 6 | 301 | 83 | 105 | -0.030 | -0.079 | 0.293 | 0.023 | 0.118 |
|---|
| MGMT |   | 5 | 360 | 366 | 379 | 0.184 | -0.022 | 0.201 | undef | 0.196 |
|---|
| XK |   | 18 | 86 | 179 | 187 | 0.154 | -0.000 | 0.067 | -0.123 | -0.040 |
|---|
| BAIAP2 |   | 3 | 557 | 412 | 430 | -0.077 | -0.025 | 0.099 | 0.092 | -0.051 |
|---|
| NR2F1 |   | 25 | 59 | 83 | 92 | -0.252 | 0.113 | 0.133 | -0.116 | 0.207 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| BCAR3 |   | 2 | 743 | 2111 | 2085 | 0.271 | 0.123 | 0.259 | -0.027 | 0.066 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| NEK1 |   | 15 | 111 | 141 | 139 | 0.032 | 0.184 | 0.049 | undef | 0.135 |
|---|
| POLE2 |   | 4 | 440 | 727 | 723 | 0.200 | -0.203 | 0.049 | 0.000 | -0.010 |
|---|
| CDC27 |   | 6 | 301 | 1010 | 986 | 0.156 | -0.086 | -0.029 | 0.006 | 0.348 |
|---|
| SH2B2 |   | 2 | 743 | 1010 | 1007 | -0.006 | -0.062 | -0.069 | -0.056 | -0.111 |
|---|
| HTT |   | 14 | 117 | 141 | 140 | -0.016 | -0.015 | 0.075 | 0.092 | -0.093 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| AURKA |   | 3 | 557 | 1165 | 1151 | 0.189 | -0.176 | 0.100 | -0.051 | 0.050 |
|---|
| ATN1 |   | 7 | 256 | 412 | 414 | -0.005 | -0.093 | 0.005 | -0.185 | -0.136 |
|---|
| TIE1 |   | 18 | 86 | 366 | 357 | -0.108 | 0.005 | 0.141 | -0.108 | 0.252 |
|---|
| CDK2 |   | 31 | 43 | 179 | 174 | 0.196 | -0.096 | 0.109 | 0.001 | 0.362 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
| IGF2 |   | 14 | 117 | 570 | 553 | -0.022 | 0.172 | 0.145 | 0.087 | 0.116 |
|---|
GO Enrichment output for subnetwork 6306 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 1.168E-08 | 2.852E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.342E-08 | 1.639E-05 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.542E-08 | 1.255E-05 |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 1.717E-08 | 1.049E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.38E-08 | 1.163E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 4.018E-08 | 1.636E-05 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 5.12E-08 | 1.787E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 5.133E-08 | 1.568E-05 |
|---|
| response to insulin stimulus | GO:0032868 |  | 7.241E-08 | 1.965E-05 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 1.187E-07 | 2.901E-05 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 1.187E-07 | 2.637E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 1.581E-08 | 3.805E-05 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.913E-08 | 2.302E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.135E-08 | 1.712E-05 |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 2.194E-08 | 1.32E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 3.406E-08 | 1.639E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 5.746E-08 | 2.304E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 5.746E-08 | 1.975E-05 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 6.539E-08 | 1.967E-05 |
|---|
| response to insulin stimulus | GO:0032868 |  | 9.342E-08 | 2.497E-05 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 1.516E-07 | 3.647E-05 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 1.516E-07 | 3.316E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 3.534E-08 | 8.129E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 7.559E-08 | 8.693E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 9.996E-08 | 7.664E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.305E-07 | 7.501E-05 |
|---|
| response to insulin stimulus | GO:0032868 |  | 1.964E-07 | 9.036E-05 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 3.168E-07 | 1.214E-04 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 6.287E-07 | 2.066E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.603E-07 | 1.898E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 6.761E-07 | 1.728E-04 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 1.123E-06 | 2.583E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.434E-06 | 2.999E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of protein kinase activity | GO:0045860 |  | 2.046E-08 | 3.77E-05 |
|---|
| positive regulation of transferase activity | GO:0051347 |  | 2.564E-08 | 2.362E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 3.468E-07 | 2.13E-04 |
|---|
| regulation of cell migration | GO:0030334 |  | 7.532E-07 | 3.47E-04 |
|---|
| regulation of locomotion | GO:0040012 |  | 1.724E-06 | 6.356E-04 |
|---|
| regulation of cell motion | GO:0051270 |  | 1.724E-06 | 5.297E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 2.234E-06 | 5.883E-04 |
|---|
| activation of MAPKK activity | GO:0000186 |  | 3.845E-06 | 8.857E-04 |
|---|
| response to insulin stimulus | GO:0032868 |  | 6.927E-06 | 1.418E-03 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 8.401E-06 | 1.548E-03 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.223E-05 | 2.048E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 3.534E-08 | 8.129E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 7.559E-08 | 8.693E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 9.996E-08 | 7.664E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.305E-07 | 7.501E-05 |
|---|
| response to insulin stimulus | GO:0032868 |  | 1.964E-07 | 9.036E-05 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 3.168E-07 | 1.214E-04 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 6.287E-07 | 2.066E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.603E-07 | 1.898E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 6.761E-07 | 1.728E-04 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 1.123E-06 | 2.583E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.434E-06 | 2.999E-04 |
|---|



