Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6111
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7615 | 3.064e-03 | 4.790e-04 | 7.498e-01 | 1.100e-06 |
|---|
| Loi | 0.2251 | 8.856e-02 | 1.365e-02 | 4.479e-01 | 5.415e-04 |
|---|
| Schmidt | 0.6734 | 0.000e+00 | 0.000e+00 | 4.491e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7901 | 0.000e+00 | 0.000e+00 | 3.553e-03 | 0.000e+00 |
|---|
| Wang | 0.2591 | 3.025e-03 | 4.690e-02 | 4.104e-01 | 5.822e-05 |
|---|
Expression data for subnetwork 6111 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
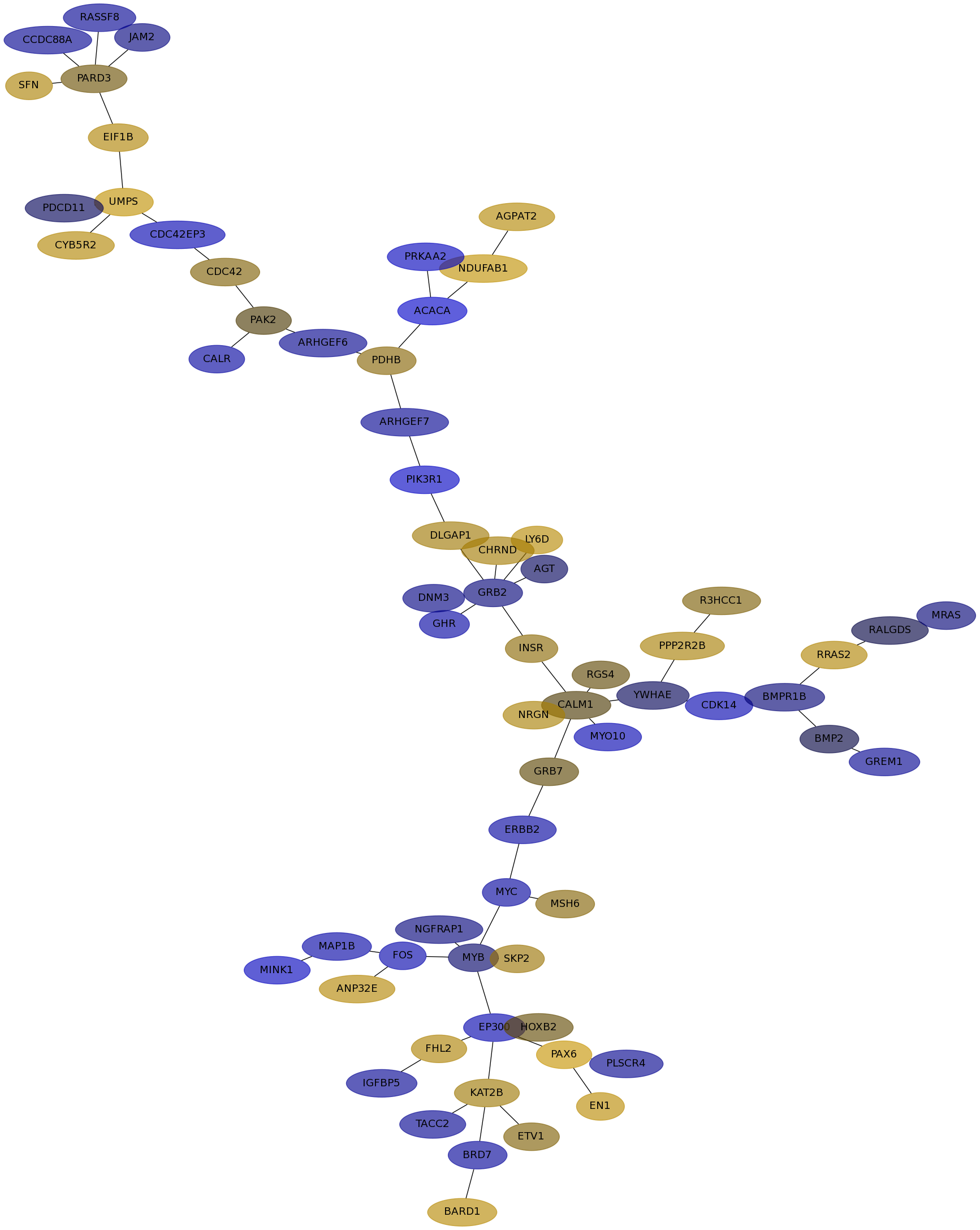
Score for each gene in subnetwork 6111 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| CDC42EP3 |   | 2 | 743 | 2284 | 2253 | -0.159 | 0.338 | -0.073 | undef | 0.220 |
|---|
| RGS4 |   | 4 | 440 | 842 | 831 | 0.022 | 0.160 | -0.006 | undef | 0.371 |
|---|
| PAX6 |   | 22 | 69 | 318 | 312 | 0.259 | -0.003 | 0.112 | undef | 0.168 |
|---|
| DNM3 |   | 1 | 1195 | 2284 | 2285 | -0.053 | -0.070 | -0.040 | undef | 0.343 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| MRAS |   | 1 | 1195 | 2284 | 2285 | -0.039 | -0.012 | 0.068 | -0.319 | -0.119 |
|---|
| NDUFAB1 |   | 2 | 743 | 1413 | 1410 | 0.230 | -0.040 | 0.162 | -0.013 | -0.051 |
|---|
| YWHAE |   | 4 | 440 | 179 | 209 | -0.018 | 0.112 | 0.279 | -0.094 | 0.014 |
|---|
| LY6D |   | 1 | 1195 | 2284 | 2285 | 0.177 | 0.008 | -0.101 | undef | 0.092 |
|---|
| EIF1B |   | 8 | 222 | 1576 | 1529 | 0.149 | 0.075 | 0.332 | -0.017 | -0.070 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| CDK14 |   | 7 | 256 | 179 | 201 | -0.151 | 0.011 | -0.071 | 0.212 | 0.107 |
|---|
| CDC42 |   | 26 | 52 | 179 | 177 | 0.047 | 0.242 | -0.027 | 0.040 | -0.042 |
|---|
| TACC2 |   | 1 | 1195 | 2284 | 2285 | -0.073 | 0.122 | 0.181 | undef | 0.180 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| ETV1 |   | 10 | 167 | 1 | 40 | 0.046 | 0.054 | -0.065 | 0.014 | 0.141 |
|---|
| ACACA |   | 3 | 557 | 1413 | 1407 | -0.275 | -0.103 | 0.056 | undef | -0.080 |
|---|
| CYB5R2 |   | 1 | 1195 | 2284 | 2285 | 0.164 | 0.230 | 0.006 | -0.041 | 0.016 |
|---|
| R3HCC1 |   | 1 | 1195 | 2284 | 2285 | 0.045 | 0.058 | 0.158 | 0.155 | -0.025 |
|---|
| PDHB |   | 13 | 124 | 525 | 503 | 0.056 | 0.017 | 0.197 | -0.205 | 0.193 |
|---|
| AGPAT2 |   | 3 | 557 | 412 | 430 | 0.167 | -0.015 | 0.123 | 0.042 | -0.076 |
|---|
| NGFRAP1 |   | 3 | 557 | 991 | 982 | -0.044 | 0.005 | 0.252 | -0.001 | 0.109 |
|---|
| ARHGEF6 |   | 10 | 167 | 525 | 507 | -0.068 | -0.120 | 0.166 | undef | -0.115 |
|---|
| ANP32E |   | 2 | 743 | 1247 | 1238 | 0.166 | -0.139 | 0.191 | undef | 0.118 |
|---|
| PDCD11 |   | 2 | 743 | 1576 | 1561 | -0.019 | 0.075 | 0.091 | undef | 0.114 |
|---|
| NRGN |   | 1 | 1195 | 2284 | 2285 | 0.131 | -0.132 | 0.141 | -0.126 | 0.004 |
|---|
| PARD3 |   | 6 | 301 | 366 | 377 | 0.030 | -0.006 | -0.052 | 0.040 | -0.090 |
|---|
| BMPR1B |   | 2 | 743 | 1709 | 1695 | -0.037 | 0.078 | 0.163 | -0.003 | 0.146 |
|---|
| PRKAA2 |   | 6 | 301 | 1165 | 1138 | -0.204 | 0.148 | 0.086 | 0.250 | -0.078 |
|---|
| MINK1 |   | 2 | 743 | 2141 | 2117 | -0.210 | -0.166 | -0.092 | 0.103 | -0.117 |
|---|
| PLSCR4 |   | 13 | 124 | 318 | 316 | -0.069 | 0.140 | 0.231 | 0.212 | 0.212 |
|---|
| KAT2B |   | 13 | 124 | 318 | 316 | 0.102 | -0.157 | -0.091 | 0.136 | -0.087 |
|---|
| CHRND |   | 3 | 557 | 318 | 341 | 0.114 | -0.031 | -0.047 | 0.154 | -0.031 |
|---|
| ARHGEF7 |   | 25 | 59 | 83 | 92 | -0.074 | -0.061 | 0.208 | 0.028 | 0.015 |
|---|
| RALGDS |   | 5 | 360 | 2024 | 1972 | -0.011 | 0.199 | 0.033 | undef | -0.009 |
|---|
| HOXB2 |   | 33 | 37 | 318 | 289 | 0.024 | -0.143 | 0.076 | 0.247 | -0.184 |
|---|
| CALR |   | 4 | 440 | 525 | 520 | -0.105 | 0.002 | -0.061 | -0.094 | -0.075 |
|---|
| BMP2 |   | 11 | 148 | 525 | 505 | -0.011 | 0.097 | 0.148 | -0.114 | -0.069 |
|---|
| RASSF8 |   | 5 | 360 | 296 | 310 | -0.078 | 0.121 | 0.174 | 0.114 | -0.092 |
|---|
| EN1 |   | 10 | 167 | 179 | 194 | 0.196 | 0.024 | 0.011 | -0.016 | 0.102 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| PAK2 |   | 26 | 52 | 179 | 177 | 0.014 | 0.161 | 0.081 | -0.044 | 0.062 |
|---|
| BRD7 |   | 23 | 66 | 457 | 437 | -0.090 | 0.019 | -0.188 | undef | -0.086 |
|---|
| JAM2 |   | 2 | 743 | 1604 | 1588 | -0.047 | 0.075 | 0.241 | -0.215 | 0.055 |
|---|
| RRAS2 |   | 7 | 256 | 1681 | 1633 | 0.155 | 0.076 | 0.289 | 0.044 | 0.031 |
|---|
| MSH6 |   | 12 | 140 | 1 | 36 | 0.052 | -0.057 | 0.051 | 0.126 | 0.064 |
|---|
| CCDC88A |   | 1 | 1195 | 2284 | 2285 | -0.072 | -0.090 | 0.097 | -0.041 | 0.019 |
|---|
| MYB |   | 28 | 47 | 366 | 344 | -0.028 | 0.075 | 0.002 | 0.065 | -0.012 |
|---|
| DLGAP1 |   | 2 | 743 | 2207 | 2182 | 0.103 | 0.078 | 0.102 | -0.072 | -0.159 |
|---|
| GREM1 |   | 9 | 196 | 525 | 508 | -0.073 | 0.200 | 0.028 | 0.225 | 0.261 |
|---|
| MYO10 |   | 3 | 557 | 570 | 572 | -0.159 | 0.121 | -0.191 | 0.074 | 0.066 |
|---|
| CALM1 |   | 6 | 301 | 614 | 602 | 0.014 | 0.021 | 0.192 | 0.221 | 0.247 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| SKP2 |   | 29 | 45 | 1 | 23 | 0.092 | -0.043 | 0.237 | undef | -0.009 |
|---|
| FHL2 |   | 21 | 73 | 658 | 630 | 0.143 | 0.239 | -0.033 | 0.107 | 0.294 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| GRB2 |   | 13 | 124 | 318 | 316 | -0.040 | -0.159 | -0.036 | -0.028 | -0.024 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| AGT |   | 4 | 440 | 1203 | 1184 | -0.020 | -0.101 | -0.043 | 0.255 | 0.064 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| UMPS |   | 3 | 557 | 842 | 834 | 0.229 | 0.047 | 0.080 | -0.098 | 0.021 |
|---|
| SFN |   | 25 | 59 | 478 | 454 | 0.141 | 0.270 | 0.023 | -0.102 | 0.101 |
|---|
GO Enrichment output for subnetwork 6111 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| filopodium assembly | GO:0046847 |  | 1.315E-06 | 3.212E-03 |
|---|
| microspike assembly | GO:0030035 |  | 1.684E-06 | 2.057E-03 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 1.824E-06 | 1.485E-03 |
|---|
| nuclear migration | GO:0007097 |  | 2.361E-06 | 1.442E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 2.361E-06 | 1.154E-03 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 4.117E-06 | 1.676E-03 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 6.563E-06 | 2.29E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 6.563E-06 | 2.004E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 6.563E-06 | 1.781E-03 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 9.208E-06 | 2.25E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 9.327E-06 | 2.071E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| filopodium assembly | GO:0046847 |  | 1.232E-06 | 2.964E-03 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 1.376E-06 | 1.655E-03 |
|---|
| microspike assembly | GO:0030035 |  | 1.604E-06 | 1.287E-03 |
|---|
| nuclear migration | GO:0007097 |  | 2.748E-06 | 1.653E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 2.748E-06 | 1.322E-03 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 4.79E-06 | 1.921E-03 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 4.79E-06 | 1.646E-03 |
|---|
| macrophage differentiation | GO:0030225 |  | 4.79E-06 | 1.441E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 7.635E-06 | 2.041E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 7.635E-06 | 1.837E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 9.716E-06 | 2.125E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| filopodium assembly | GO:0046847 |  | 6.94E-07 | 1.596E-03 |
|---|
| microspike assembly | GO:0030035 |  | 1.035E-06 | 1.191E-03 |
|---|
| nuclear migration | GO:0007097 |  | 3.202E-06 | 2.455E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.202E-06 | 1.841E-03 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 3.464E-06 | 1.593E-03 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 6.371E-06 | 2.442E-03 |
|---|
| macrophage differentiation | GO:0030225 |  | 6.371E-06 | 2.093E-03 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.109E-05 | 3.189E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.109E-05 | 2.835E-03 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.766E-05 | 4.062E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.766E-05 | 3.693E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regionalization | GO:0003002 |  | 1.75E-06 | 3.225E-03 |
|---|
| dorsal/ventral pattern formation | GO:0009953 |  | 2.89E-06 | 2.663E-03 |
|---|
| positive regulation of growth | GO:0045927 |  | 6.499E-06 | 3.993E-03 |
|---|
| positive regulation of transferase activity | GO:0051347 |  | 7.33E-06 | 3.377E-03 |
|---|
| pattern specification process | GO:0007389 |  | 1.075E-05 | 3.963E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.473E-05 | 4.526E-03 |
|---|
| hindbrain development | GO:0030902 |  | 2.488E-05 | 6.552E-03 |
|---|
| proximal/distal pattern formation | GO:0009954 |  | 2.488E-05 | 5.733E-03 |
|---|
| negative regulation of translation | GO:0017148 |  | 3.305E-05 | 6.768E-03 |
|---|
| regulation of cell migration | GO:0030334 |  | 3.91E-05 | 7.207E-03 |
|---|
| negative regulation of signal transduction | GO:0009968 |  | 3.91E-05 | 6.552E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| filopodium assembly | GO:0046847 |  | 6.94E-07 | 1.596E-03 |
|---|
| microspike assembly | GO:0030035 |  | 1.035E-06 | 1.191E-03 |
|---|
| nuclear migration | GO:0007097 |  | 3.202E-06 | 2.455E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.202E-06 | 1.841E-03 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 3.464E-06 | 1.593E-03 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 6.371E-06 | 2.442E-03 |
|---|
| macrophage differentiation | GO:0030225 |  | 6.371E-06 | 2.093E-03 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.109E-05 | 3.189E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.109E-05 | 2.835E-03 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.766E-05 | 4.062E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.766E-05 | 3.693E-03 |
|---|



