Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 5717-7-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2348 | 6.380e-03 | 7.600e-03 | 7.044e-02 |
|---|
| IPC-NIBC-129 | 0.2789 | 3.497e-02 | 4.346e-02 | 3.521e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2700 | 3.851e-03 | 6.526e-03 | 1.432e-01 |
|---|
| Loi_GPL570 | 0.2874 | 4.133e-02 | 4.101e-02 | 2.210e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1751 | 2.500e-05 | 1.450e-04 | 2.970e-04 |
|---|
| Parker_GPL1390 | 0.2841 | 6.025e-02 | 5.732e-02 | 4.174e-01 |
|---|
| Parker_GPL887 | 0.0537 | 8.259e-01 | 8.429e-01 | 8.780e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3423 | 4.182e-01 | 3.814e-01 | 9.402e-01 |
|---|
| Schmidt | 0.2261 | 9.542e-03 | 1.366e-02 | 1.261e-01 |
|---|
| Sotiriou | 0.3180 | 4.508e-03 | 1.522e-03 | 9.473e-02 |
|---|
| Wang | 0.1891 | 8.683e-02 | 5.831e-02 | 3.330e-01 |
|---|
| Zhang | 0.1248 | 9.187e-02 | 5.917e-02 | 2.229e-01 |
|---|
| Zhou | 0.3773 | 7.257e-02 | 7.796e-02 | 4.352e-01 |
|---|
Expression data for subnetwork 5717-7-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
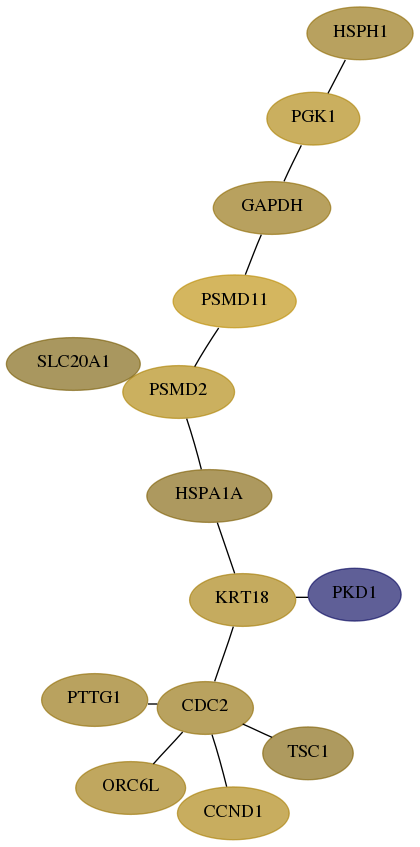
Score for each gene in subnetwork 5717-7-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| psmd2 |   | 7 | 38 | 37 | 28 | 0.147 | 0.322 | 0.122 | 0.146 | 0.143 | 0.246 | 0.444 | 0.194 | 0.318 | 0.151 | 0.154 | 0.042 | 0.207 |
|---|
| pttg1 |   | 10 | 25 | 31 | 21 | 0.072 | 0.270 | 0.175 | 0.183 | 0.200 | 0.208 | 0.347 | 0.334 | 0.224 | 0.204 | 0.147 | 0.210 | 0.309 |
|---|
| krt18 |   | 15 | 15 | 230 | 195 | 0.112 | -0.078 | 0.147 | 0.150 | 0.161 | -0.086 | -0.204 | 0.083 | -0.099 | 0.118 | 0.086 | -0.114 | 0.222 |
|---|
| pgk1 |   | 5 | 50 | 53 | 47 | 0.141 | 0.293 | 0.135 | 0.197 | 0.170 | 0.170 | 0.120 | 0.310 | 0.201 | 0.127 | 0.110 | 0.054 | 0.141 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| tsc1 |   | 47 | 5 | 17 | 8 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.043 | -0.007 | 0.207 |
|---|
| psmd11 |   | 1 | 164 | 312 | 318 | 0.200 | 0.177 | 0.125 | 0.068 | 0.122 | 0.121 | -0.033 | 0.134 | 0.137 | 0.074 | 0.118 | -0.075 | 0.191 |
|---|
| pkd1 |   | 9 | 27 | 234 | 205 | -0.021 | 0.055 | 0.007 | 0.037 | 0.071 | 0.184 | 0.256 | -0.217 | -0.068 | -0.001 | 0.046 | 0.077 | 0.151 |
|---|
| orc6l |   | 8 | 33 | 73 | 61 | 0.094 | 0.270 | 0.084 | 0.114 | 0.106 | 0.240 | 0.256 | 0.234 | 0.192 | 0.105 | 0.086 | 0.175 | 0.160 |
|---|
| gapdh |   | 6 | 43 | 136 | 117 | 0.071 | 0.312 | 0.102 | 0.145 | 0.165 | 0.172 | -0.253 | 0.288 | 0.064 | 0.213 | 0.056 | -0.088 | 0.235 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| hsph1 |   | 4 | 60 | 53 | 50 | 0.071 | 0.260 | 0.138 | 0.050 | 0.148 | 0.197 | -0.116 | 0.161 | 0.143 | 0.132 | 0.169 | 0.094 | 0.357 |
|---|
| slc20a1 |   | 7 | 38 | 37 | 28 | 0.041 | 0.039 | 0.109 | 0.170 | 0.101 | 0.121 | 0.326 | 0.086 | 0.048 | 0.124 | 0.121 | -0.050 | 0.299 |
|---|
| hspa1a |   | 4 | 60 | 37 | 39 | 0.047 | -0.046 | 0.070 | 0.019 | 0.160 | 0.104 | 0.230 | 0.108 | 0.121 | 0.110 | 0.175 | -0.055 | 0.192 |
|---|
GO Enrichment output for subnetwork 5717-7-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 1.743E-11 | 4.008E-08 |
|---|
| glucose catabolic process | GO:0006007 |  | 6.323E-11 | 7.272E-08 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.82E-10 | 1.396E-07 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 3.457E-10 | 1.988E-07 |
|---|
| alcohol catabolic process | GO:0046164 |  | 7.199E-10 | 3.311E-07 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.806E-09 | 6.924E-07 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.986E-08 | 6.525E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.662E-07 | 4.779E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.13E-06 | 2.889E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.207E-06 | 2.777E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.461E-06 | 3.055E-04 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 9.705E-13 | 2.371E-09 |
|---|
| glucose catabolic process | GO:0006007 |  | 3.347E-12 | 4.089E-09 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.122E-11 | 9.133E-09 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 2.418E-11 | 1.477E-08 |
|---|
| alcohol catabolic process | GO:0046164 |  | 5.125E-11 | 2.504E-08 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 3.905E-09 | 1.59E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 6.423E-08 | 2.242E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.079E-07 | 6.35E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 2.206E-07 | 5.989E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 2.621E-07 | 6.404E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 2.621E-07 | 5.822E-05 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 1.77E-12 | 4.26E-09 |
|---|
| glucose catabolic process | GO:0006007 |  | 6.266E-12 | 7.537E-09 |
|---|
| hexose catabolic process | GO:0019320 |  | 2.142E-11 | 1.718E-08 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 4.669E-11 | 2.808E-08 |
|---|
| alcohol catabolic process | GO:0046164 |  | 9.983E-11 | 4.804E-08 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 5.705E-09 | 2.288E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 9.381E-08 | 3.224E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.422E-07 | 1.029E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.63E-07 | 9.705E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 4.313E-07 | 1.038E-04 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 4.313E-07 | 9.433E-05 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 9.705E-13 | 2.371E-09 |
|---|
| glucose catabolic process | GO:0006007 |  | 3.347E-12 | 4.089E-09 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.122E-11 | 9.133E-09 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 2.418E-11 | 1.477E-08 |
|---|
| alcohol catabolic process | GO:0046164 |  | 5.125E-11 | 2.504E-08 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 3.905E-09 | 1.59E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 6.423E-08 | 2.242E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 2.079E-07 | 6.35E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 2.206E-07 | 5.989E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 2.621E-07 | 6.404E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 2.621E-07 | 5.822E-05 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 1.77E-12 | 4.26E-09 |
|---|
| glucose catabolic process | GO:0006007 |  | 6.266E-12 | 7.537E-09 |
|---|
| hexose catabolic process | GO:0019320 |  | 2.142E-11 | 1.718E-08 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 4.669E-11 | 2.808E-08 |
|---|
| alcohol catabolic process | GO:0046164 |  | 9.983E-11 | 4.804E-08 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 5.705E-09 | 2.288E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 9.381E-08 | 3.224E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.422E-07 | 1.029E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.63E-07 | 9.705E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 4.313E-07 | 1.038E-04 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 4.313E-07 | 9.433E-05 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of Ras GTPase activity | GO:0032318 |  | 7.739E-07 | 2.764E-03 |
|---|
| Ras GTPase activator activity | GO:0005099 |  | 9.421E-07 | 1.682E-03 |
|---|
| regulation of GTPase activity | GO:0043087 |  | 1.68E-06 | 1.999E-03 |
|---|
| cysteine-type endopeptidase inhibitor activity | GO:0004869 |  | 2.731E-06 | 2.438E-03 |
|---|
| regulation of Rab protein signal transduction | GO:0032483 |  | 6.609E-06 | 4.72E-03 |
|---|
| inorganic anion transmembrane transporter activity | GO:0015103 |  | 6.609E-06 | 3.933E-03 |
|---|
| Rab GTPase activator activity | GO:0005097 |  | 7.127E-06 | 3.636E-03 |
|---|
| glycolysis | GO:0006096 |  | 7.671E-06 | 3.424E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 8.371E-06 | 3.321E-03 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 8.371E-06 | 2.989E-03 |
|---|
| proteasome complex | GO:0000502 |  | 1.08E-05 | 3.507E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.927E-07 | 7.028E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 2.335E-07 | 4.258E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 2.64E-07 | 3.211E-04 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 2.804E-07 | 2.557E-04 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 3.154E-07 | 2.301E-04 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 4.17E-07 | 2.535E-04 |
|---|
| regulation of ligase activity | GO:0051340 |  | 4.886E-07 | 2.546E-04 |
|---|
| proteasomal ubiquitin-dependent protein catabolic process | GO:0043161 |  | 6.278E-07 | 2.863E-04 |
|---|
| glycolysis | GO:0006096 |  | 5.354E-06 | 2.17E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 6.839E-06 | 2.495E-03 |
|---|
| origin recognition complex | GO:0000808 |  | 6.839E-06 | 2.268E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 1.77E-12 | 4.26E-09 |
|---|
| glucose catabolic process | GO:0006007 |  | 6.266E-12 | 7.537E-09 |
|---|
| hexose catabolic process | GO:0019320 |  | 2.142E-11 | 1.718E-08 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 4.669E-11 | 2.808E-08 |
|---|
| alcohol catabolic process | GO:0046164 |  | 9.983E-11 | 4.804E-08 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 5.705E-09 | 2.288E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 9.381E-08 | 3.224E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 3.422E-07 | 1.029E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 3.63E-07 | 9.705E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 4.313E-07 | 1.038E-04 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 4.313E-07 | 9.433E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 1.743E-11 | 4.008E-08 |
|---|
| glucose catabolic process | GO:0006007 |  | 6.323E-11 | 7.272E-08 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.82E-10 | 1.396E-07 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 3.457E-10 | 1.988E-07 |
|---|
| alcohol catabolic process | GO:0046164 |  | 7.199E-10 | 3.311E-07 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.806E-09 | 6.924E-07 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.986E-08 | 6.525E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.662E-07 | 4.779E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.13E-06 | 2.889E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.207E-06 | 2.777E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.461E-06 | 3.055E-04 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 1.743E-11 | 4.008E-08 |
|---|
| glucose catabolic process | GO:0006007 |  | 6.323E-11 | 7.272E-08 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.82E-10 | 1.396E-07 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 3.457E-10 | 1.988E-07 |
|---|
| alcohol catabolic process | GO:0046164 |  | 7.199E-10 | 3.311E-07 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.806E-09 | 6.924E-07 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.986E-08 | 6.525E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.662E-07 | 4.779E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.13E-06 | 2.889E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.207E-06 | 2.777E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.461E-06 | 3.055E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 1.743E-11 | 4.008E-08 |
|---|
| glucose catabolic process | GO:0006007 |  | 6.323E-11 | 7.272E-08 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.82E-10 | 1.396E-07 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 3.457E-10 | 1.988E-07 |
|---|
| alcohol catabolic process | GO:0046164 |  | 7.199E-10 | 3.311E-07 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.806E-09 | 6.924E-07 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.986E-08 | 6.525E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.662E-07 | 4.779E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.13E-06 | 2.889E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.207E-06 | 2.777E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.461E-06 | 3.055E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 1.743E-11 | 4.008E-08 |
|---|
| glucose catabolic process | GO:0006007 |  | 6.323E-11 | 7.272E-08 |
|---|
| hexose catabolic process | GO:0019320 |  | 1.82E-10 | 1.396E-07 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 3.457E-10 | 1.988E-07 |
|---|
| alcohol catabolic process | GO:0046164 |  | 7.199E-10 | 3.311E-07 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 1.806E-09 | 6.924E-07 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.986E-08 | 6.525E-06 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 1.662E-07 | 4.779E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.13E-06 | 2.889E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.207E-06 | 2.777E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.461E-06 | 3.055E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycolysis | GO:0006096 |  | 2.237E-13 | 5.307E-10 |
|---|
| glucose catabolic process | GO:0006007 |  | 9.963E-13 | 1.182E-09 |
|---|
| hexose catabolic process | GO:0019320 |  | 3.067E-12 | 2.426E-09 |
|---|
| monosaccharide catabolic process | GO:0046365 |  | 6.116E-12 | 3.628E-09 |
|---|
| alcohol catabolic process | GO:0046164 |  | 1.356E-11 | 6.437E-09 |
|---|
| carbohydrate catabolic process | GO:0016052 |  | 3.993E-11 | 1.579E-08 |
|---|
| detection of mechanical stimulus | GO:0050982 |  | 1.627E-09 | 5.516E-07 |
|---|
| cartilage condensation | GO:0001502 |  | 2.276E-09 | 6.752E-07 |
|---|
| substrate-bound cell migration. cell extension | GO:0006930 |  | 1.98E-08 | 5.221E-06 |
|---|
| response to mechanical stimulus | GO:0009612 |  | 1.036E-07 | 2.459E-05 |
|---|
| substrate-bound cell migration | GO:0006929 |  | 2.366E-07 | 5.103E-05 |
|---|



