Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 103-0-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2626 | 4.456e-03 | 5.290e-03 | 5.330e-02 |
|---|
| IPC-NIBC-129 | 0.2441 | 3.035e-02 | 3.731e-02 | 3.164e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2874 | 3.487e-03 | 5.969e-03 | 1.339e-01 |
|---|
| Loi_GPL570 | 0.2920 | 6.662e-02 | 6.617e-02 | 3.136e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1342 | 2.400e-05 | 1.400e-04 | 2.800e-04 |
|---|
| Parker_GPL1390 | 0.2943 | 4.053e-02 | 3.707e-02 | 3.347e-01 |
|---|
| Parker_GPL887 | 0.1150 | 5.963e-01 | 6.269e-01 | 6.439e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3860 | 7.094e-01 | 6.938e-01 | 9.961e-01 |
|---|
| Schmidt | 0.2267 | 4.736e-03 | 6.981e-03 | 7.189e-02 |
|---|
| Sotiriou | 0.3606 | 2.812e-03 | 8.390e-04 | 6.783e-02 |
|---|
| Wang | 0.2094 | 1.669e-01 | 1.251e-01 | 5.032e-01 |
|---|
| Zhang | 0.0621 | 6.853e-02 | 4.166e-02 | 1.707e-01 |
|---|
| Zhou | 0.4058 | 4.780e-02 | 4.933e-02 | 3.044e-01 |
|---|
Expression data for subnetwork 103-0-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
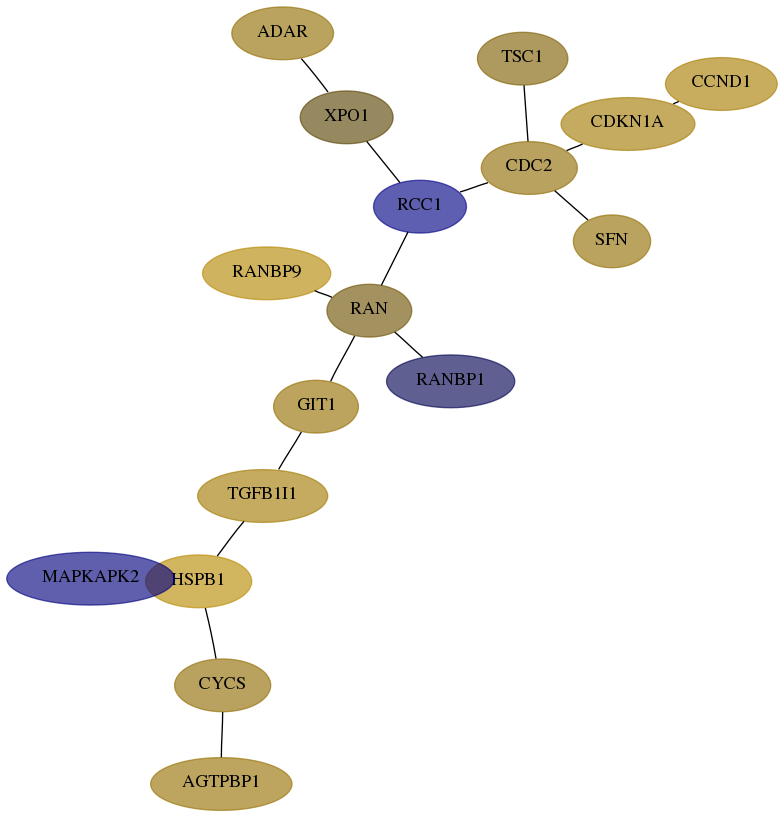
Score for each gene in subnetwork 103-0-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| agtpbp1 |   | 11 | 21 | 17 | 13 | 0.086 | 0.219 | 0.009 | 0.181 | 0.071 | 0.078 | 0.296 | 0.026 | 0.110 | 0.151 | 0.103 | 0.064 | 0.226 |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| ran |   | 4 | 60 | 100 | 91 | 0.032 | 0.181 | 0.147 | 0.083 | 0.166 | 0.250 | -0.287 | 0.247 | 0.127 | 0.178 | 0.052 | -0.025 | 0.254 |
|---|
| adar |   | 1 | 164 | 174 | 184 | 0.078 | 0.097 | 0.022 | -0.181 | 0.079 | -0.163 | -0.137 | 0.154 | 0.006 | 0.060 | -0.037 | 0.113 | 0.213 |
|---|
| tsc1 |   | 47 | 5 | 17 | 8 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.043 | -0.007 | 0.207 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| ranbp9 |   | 4 | 60 | 100 | 91 | 0.172 | 0.207 | 0.012 | 0.253 | 0.126 | 0.186 | -0.113 | 0.148 | 0.143 | 0.127 | 0.205 | 0.021 | 0.164 |
|---|
| rcc1 |   | 4 | 60 | 100 | 91 | -0.056 | 0.114 | 0.136 | 0.093 | 0.114 | 0.146 | 0.325 | 0.167 | 0.113 | 0.063 | -0.108 | 0.072 | 0.253 |
|---|
| git1 |   | 5 | 50 | 78 | 70 | 0.082 | -0.027 | 0.097 | 0.051 | 0.077 | 0.074 | -0.099 | 0.138 | 0.087 | 0.097 | -0.065 | -0.065 | 0.114 |
|---|
| cycs |   | 10 | 25 | 17 | 14 | 0.073 | 0.206 | 0.080 | 0.094 | 0.117 | -0.056 | 0.202 | 0.272 | 0.157 | 0.077 | 0.190 | 0.135 | 0.234 |
|---|
| ranbp1 |   | 4 | 60 | 100 | 91 | -0.016 | 0.220 | 0.154 | 0.205 | 0.114 | 0.234 | 0.180 | 0.253 | 0.217 | 0.082 | -0.006 | -0.003 | 0.136 |
|---|
| hspb1 |   | 19 | 11 | 17 | 11 | 0.190 | -0.050 | 0.184 | 0.212 | 0.231 | -0.026 | -0.065 | 0.289 | -0.021 | 0.201 | 0.125 | -0.077 | 0.290 |
|---|
| xpo1 |   | 3 | 79 | 100 | 95 | 0.020 | 0.153 | 0.081 | 0.020 | 0.092 | 0.151 | -0.080 | 0.101 | 0.194 | 0.104 | 0.140 | 0.090 | 0.317 |
|---|
| mapkapk2 |   | 13 | 19 | 17 | 12 | -0.050 | 0.102 | 0.156 | -0.116 | 0.090 | 0.168 | 0.303 | 0.074 | 0.171 | 0.117 | 0.085 | -0.124 | 0.312 |
|---|
| tgfb1i1 |   | 3 | 79 | 100 | 95 | 0.114 | 0.089 | -0.023 | -0.034 | -0.008 | 0.152 | 0.381 | -0.183 | -0.102 | -0.076 | 0.198 | -0.065 | -0.057 |
|---|
| sfn |   | 50 | 4 | 29 | 17 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.025 | 0.079 | 0.181 |
|---|
GO Enrichment output for subnetwork 103-0-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle organization | GO:0007051 |  | 1.402E-07 | 3.224E-04 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 2.046E-07 | 2.352E-04 |
|---|
| mitotic spindle organization | GO:0007052 |  | 6.019E-07 | 4.615E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.222E-07 | 3.578E-04 |
|---|
| nuclear export | GO:0051168 |  | 9.757E-07 | 4.488E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.249E-06 | 4.788E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 2.032E-06 | 6.677E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.021E-05 | 2.936E-03 |
|---|
| interphase | GO:0051325 |  | 1.119E-05 | 2.86E-03 |
|---|
| establishment of RNA localization | GO:0051236 |  | 1.119E-05 | 2.574E-03 |
|---|
| RNA localization | GO:0006403 |  | 1.278E-05 | 2.673E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle organization | GO:0007051 |  | 3.907E-08 | 9.545E-05 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 6.289E-08 | 7.683E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.754E-07 | 1.429E-04 |
|---|
| mitotic spindle organization | GO:0007052 |  | 2.464E-07 | 1.505E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.211E-07 | 2.057E-04 |
|---|
| nuclear export | GO:0051168 |  | 4.844E-07 | 1.972E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 7.587E-07 | 2.648E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 1.079E-06 | 3.296E-04 |
|---|
| regulation of caspase activity | GO:0043281 |  | 1.211E-06 | 3.286E-04 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 1.421E-06 | 3.471E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.479E-06 | 3.286E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle organization | GO:0007051 |  | 6.762E-08 | 1.627E-04 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 9.518E-08 | 1.145E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.031E-07 | 2.431E-04 |
|---|
| mitotic spindle organization | GO:0007052 |  | 3.726E-07 | 2.241E-04 |
|---|
| nuclear export | GO:0051168 |  | 6.285E-07 | 3.024E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.266E-07 | 2.914E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.147E-06 | 3.942E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 1.632E-06 | 4.907E-04 |
|---|
| regulation of caspase activity | GO:0043281 |  | 1.868E-06 | 4.993E-04 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 2.085E-06 | 5.017E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 2.236E-06 | 4.89E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle organization | GO:0007051 |  | 3.907E-08 | 9.545E-05 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 6.289E-08 | 7.683E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.754E-07 | 1.429E-04 |
|---|
| mitotic spindle organization | GO:0007052 |  | 2.464E-07 | 1.505E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.211E-07 | 2.057E-04 |
|---|
| nuclear export | GO:0051168 |  | 4.844E-07 | 1.972E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 7.587E-07 | 2.648E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 1.079E-06 | 3.296E-04 |
|---|
| regulation of caspase activity | GO:0043281 |  | 1.211E-06 | 3.286E-04 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 1.421E-06 | 3.471E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.479E-06 | 3.286E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle organization | GO:0007051 |  | 6.762E-08 | 1.627E-04 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 9.518E-08 | 1.145E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.031E-07 | 2.431E-04 |
|---|
| mitotic spindle organization | GO:0007052 |  | 3.726E-07 | 2.241E-04 |
|---|
| nuclear export | GO:0051168 |  | 6.285E-07 | 3.024E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.266E-07 | 2.914E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.147E-06 | 3.942E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 1.632E-06 | 4.907E-04 |
|---|
| regulation of caspase activity | GO:0043281 |  | 1.868E-06 | 4.993E-04 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 2.085E-06 | 5.017E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 2.236E-06 | 4.89E-04 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle organization | GO:0007051 |  | 2.167E-08 | 7.737E-05 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 2.652E-08 | 4.735E-05 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 5.864E-08 | 6.98E-05 |
|---|
| nuclear export | GO:0051168 |  | 8.277E-08 | 7.389E-05 |
|---|
| Ran GTPase binding | GO:0008536 |  | 1.255E-07 | 8.963E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.472E-07 | 8.762E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.724E-07 | 8.797E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.072E-07 | 9.25E-05 |
|---|
| mitotic spindle organization | GO:0007052 |  | 2.298E-07 | 9.117E-05 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 3.797E-07 | 1.356E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 4.743E-07 | 1.54E-04 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle organization | GO:0007051 |  | 1.31E-08 | 4.78E-05 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 2.132E-08 | 3.889E-05 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 4.948E-08 | 6.017E-05 |
|---|
| Ran GTPase binding | GO:0008536 |  | 7.417E-08 | 6.764E-05 |
|---|
| nuclear export | GO:0051168 |  | 8.277E-08 | 6.039E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.12E-08 | 5.545E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.837E-07 | 9.575E-05 |
|---|
| mitotic spindle organization | GO:0007052 |  | 1.939E-07 | 8.841E-05 |
|---|
| protein export from nucleus | GO:0006611 |  | 3.204E-07 | 1.299E-04 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 3.204E-07 | 1.169E-04 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 4.923E-07 | 1.633E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle organization | GO:0007051 |  | 6.762E-08 | 1.627E-04 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 9.518E-08 | 1.145E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.031E-07 | 2.431E-04 |
|---|
| mitotic spindle organization | GO:0007052 |  | 3.726E-07 | 2.241E-04 |
|---|
| nuclear export | GO:0051168 |  | 6.285E-07 | 3.024E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.266E-07 | 2.914E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.147E-06 | 3.942E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 1.632E-06 | 4.907E-04 |
|---|
| regulation of caspase activity | GO:0043281 |  | 1.868E-06 | 4.993E-04 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 2.085E-06 | 5.017E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 2.236E-06 | 4.89E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle organization | GO:0007051 |  | 1.402E-07 | 3.224E-04 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 2.046E-07 | 2.352E-04 |
|---|
| mitotic spindle organization | GO:0007052 |  | 6.019E-07 | 4.615E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.222E-07 | 3.578E-04 |
|---|
| nuclear export | GO:0051168 |  | 9.757E-07 | 4.488E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.249E-06 | 4.788E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 2.032E-06 | 6.677E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.021E-05 | 2.936E-03 |
|---|
| interphase | GO:0051325 |  | 1.119E-05 | 2.86E-03 |
|---|
| establishment of RNA localization | GO:0051236 |  | 1.119E-05 | 2.574E-03 |
|---|
| RNA localization | GO:0006403 |  | 1.278E-05 | 2.673E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle organization | GO:0007051 |  | 1.402E-07 | 3.224E-04 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 2.046E-07 | 2.352E-04 |
|---|
| mitotic spindle organization | GO:0007052 |  | 6.019E-07 | 4.615E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.222E-07 | 3.578E-04 |
|---|
| nuclear export | GO:0051168 |  | 9.757E-07 | 4.488E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.249E-06 | 4.788E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 2.032E-06 | 6.677E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.021E-05 | 2.936E-03 |
|---|
| interphase | GO:0051325 |  | 1.119E-05 | 2.86E-03 |
|---|
| establishment of RNA localization | GO:0051236 |  | 1.119E-05 | 2.574E-03 |
|---|
| RNA localization | GO:0006403 |  | 1.278E-05 | 2.673E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle organization | GO:0007051 |  | 1.402E-07 | 3.224E-04 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 2.046E-07 | 2.352E-04 |
|---|
| mitotic spindle organization | GO:0007052 |  | 6.019E-07 | 4.615E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.222E-07 | 3.578E-04 |
|---|
| nuclear export | GO:0051168 |  | 9.757E-07 | 4.488E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.249E-06 | 4.788E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 2.032E-06 | 6.677E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.021E-05 | 2.936E-03 |
|---|
| interphase | GO:0051325 |  | 1.119E-05 | 2.86E-03 |
|---|
| establishment of RNA localization | GO:0051236 |  | 1.119E-05 | 2.574E-03 |
|---|
| RNA localization | GO:0006403 |  | 1.278E-05 | 2.673E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle organization | GO:0007051 |  | 1.402E-07 | 3.224E-04 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 2.046E-07 | 2.352E-04 |
|---|
| mitotic spindle organization | GO:0007052 |  | 6.019E-07 | 4.615E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.222E-07 | 3.578E-04 |
|---|
| nuclear export | GO:0051168 |  | 9.757E-07 | 4.488E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.249E-06 | 4.788E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 2.032E-06 | 6.677E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.021E-05 | 2.936E-03 |
|---|
| interphase | GO:0051325 |  | 1.119E-05 | 2.86E-03 |
|---|
| establishment of RNA localization | GO:0051236 |  | 1.119E-05 | 2.574E-03 |
|---|
| RNA localization | GO:0006403 |  | 1.278E-05 | 2.673E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle organization | GO:0007051 |  | 1.25E-07 | 2.967E-04 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 1.535E-07 | 1.822E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.104E-07 | 3.246E-04 |
|---|
| mitotic spindle organization | GO:0007052 |  | 5.452E-07 | 3.234E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.775E-07 | 4.165E-04 |
|---|
| nuclear export | GO:0051168 |  | 1.179E-06 | 4.664E-04 |
|---|
| protein export from nucleus | GO:0006611 |  | 1.677E-06 | 5.685E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.455E-06 | 1.915E-03 |
|---|
| interphase | GO:0051325 |  | 7.067E-06 | 1.863E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 1.881E-05 | 4.463E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.906E-05 | 4.113E-03 |
|---|



