Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 941
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3395 | 1.139e-03 | 1.700e-05 | 1.437e-01 | 2.783e-09 |
|---|
| IPC | 0.3568 | 3.225e-01 | 1.560e-01 | 9.301e-01 | 4.680e-02 |
|---|
| Loi | 0.4500 | 1.810e-04 | 0.000e+00 | 6.800e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5823 | 2.000e-06 | 0.000e+00 | 1.533e-01 | 0.000e+00 |
|---|
| Wang | 0.2527 | 9.172e-03 | 7.430e-02 | 2.500e-01 | 1.703e-04 |
|---|
Expression data for subnetwork 941 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
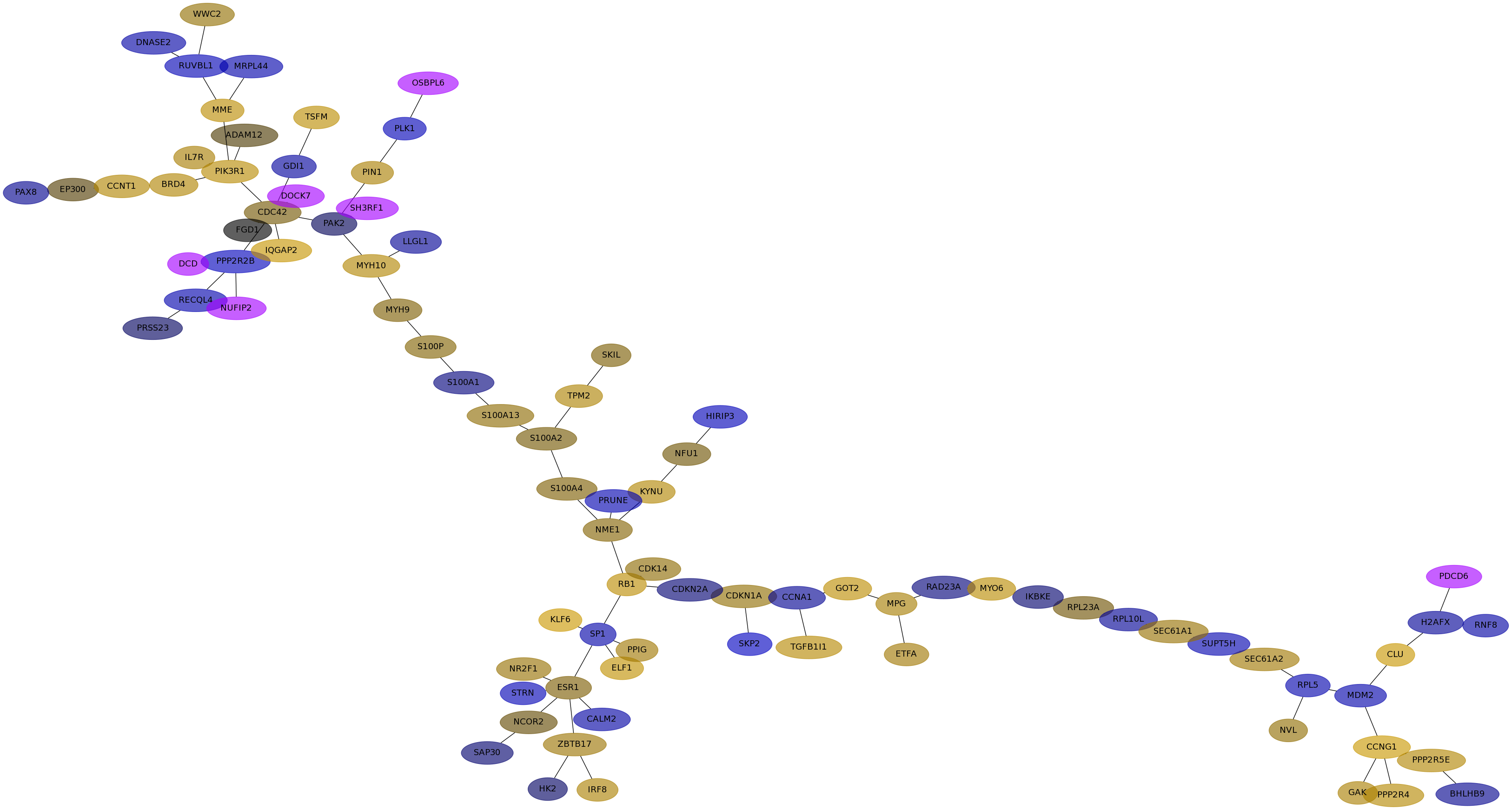
Score for each gene in subnetwork 941 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| TSFM |   | 1 | 652 | 1356 | 1357 | 0.203 | 0.155 | -0.002 | 0.065 | 0.076 |
|---|
| MYH9 |   | 2 | 391 | 1319 | 1320 | 0.047 | -0.103 | 0.227 | 0.007 | 0.165 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| NUFIP2 |   | 1 | 652 | 1356 | 1357 | undef | -0.097 | -0.167 | undef | undef |
|---|
| MYH10 |   | 2 | 391 | 1236 | 1221 | 0.163 | -0.029 | 0.139 | 0.132 | 0.223 |
|---|
| RAD23A |   | 1 | 652 | 1356 | 1357 | -0.044 | -0.013 | 0.056 | -0.166 | 0.031 |
|---|
| CDK14 |   | 2 | 391 | 1356 | 1337 | 0.067 | -0.151 | 0.011 | -0.071 | 0.107 |
|---|
| RPL23A |   | 10 | 67 | 57 | 52 | 0.031 | 0.208 | -0.007 | 0.024 | 0.055 |
|---|
| CALM2 |   | 1 | 652 | 1356 | 1357 | -0.114 | 0.076 | 0.056 | 0.104 | 0.086 |
|---|
| STRN |   | 1 | 652 | 1356 | 1357 | -0.174 | -0.044 | 0.147 | -0.054 | -0.118 |
|---|
| MDM2 |   | 1 | 652 | 1356 | 1357 | -0.137 | -0.110 | -0.038 | 0.113 | -0.108 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| NVL |   | 1 | 652 | 1356 | 1357 | 0.075 | 0.087 | 0.123 | -0.077 | 0.138 |
|---|
| GOT2 |   | 2 | 391 | 1052 | 1044 | 0.210 | -0.061 | 0.100 | -0.238 | 0.105 |
|---|
| DCD |   | 2 | 391 | 57 | 80 | undef | 0.135 | undef | undef | undef |
|---|
| PIN1 |   | 17 | 30 | 1 | 13 | 0.135 | 0.194 | 0.040 | -0.040 | -0.048 |
|---|
| S100P |   | 1 | 652 | 1356 | 1357 | 0.053 | 0.261 | 0.192 | -0.154 | 0.018 |
|---|
| ETFA |   | 3 | 265 | 756 | 748 | 0.105 | 0.027 | 0.094 | 0.088 | 0.013 |
|---|
| HIRIP3 |   | 1 | 652 | 1356 | 1357 | -0.193 | 0.167 | -0.099 | -0.068 | 0.059 |
|---|
| ELF1 |   | 2 | 391 | 1070 | 1062 | 0.229 | -0.076 | 0.083 | -0.101 | -0.077 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| NFU1 |   | 2 | 391 | 963 | 966 | 0.032 | 0.280 | 0.023 | 0.085 | -0.257 |
|---|
| RPL10L |   | 1 | 652 | 1356 | 1357 | -0.084 | 0.056 | -0.083 | 0.069 | 0.084 |
|---|
| WWC2 |   | 1 | 652 | 1356 | 1357 | 0.079 | -0.177 | 0.303 | -0.086 | 0.029 |
|---|
| S100A13 |   | 1 | 652 | 1356 | 1357 | 0.068 | 0.286 | 0.092 | 0.019 | 0.045 |
|---|
| SEC61A2 |   | 3 | 265 | 271 | 280 | 0.105 | -0.035 | -0.072 | -0.115 | -0.073 |
|---|
| SAP30 |   | 2 | 391 | 806 | 806 | -0.033 | 0.186 | 0.083 | 0.153 | -0.184 |
|---|
| ZBTB17 |   | 2 | 391 | 937 | 928 | 0.094 | 0.067 | 0.256 | -0.145 | 0.255 |
|---|
| PLK1 |   | 5 | 148 | 444 | 440 | -0.179 | 0.142 | -0.143 | 0.094 | -0.163 |
|---|
| BRD4 |   | 18 | 26 | 1 | 12 | 0.156 | -0.119 | 0.095 | -0.075 | -0.044 |
|---|
| S100A4 |   | 6 | 120 | 283 | 282 | 0.047 | 0.163 | -0.116 | 0.096 | 0.148 |
|---|
| SKIL |   | 2 | 391 | 1336 | 1322 | 0.045 | 0.209 | 0.108 | 0.073 | -0.020 |
|---|
| PPP2R5E |   | 3 | 265 | 816 | 804 | 0.160 | 0.001 | 0.191 | 0.018 | 0.094 |
|---|
| RUVBL1 |   | 1 | 652 | 1356 | 1357 | -0.173 | 0.112 | -0.107 | 0.049 | 0.027 |
|---|
| SEC61A1 |   | 5 | 148 | 271 | 267 | 0.085 | -0.161 | 0.128 | -0.096 | 0.070 |
|---|
| NME1 |   | 7 | 100 | 492 | 495 | 0.054 | 0.267 | 0.106 | 0.046 | -0.035 |
|---|
| CCNT1 |   | 18 | 26 | 33 | 32 | 0.163 | 0.008 | -0.028 | 0.162 | -0.213 |
|---|
| SH3RF1 |   | 14 | 42 | 117 | 120 | undef | 0.144 | 0.238 | undef | undef |
|---|
| BHLHB9 |   | 1 | 652 | 1356 | 1357 | -0.070 | -0.077 | 0.016 | -0.010 | 0.039 |
|---|
| KYNU |   | 4 | 198 | 606 | 597 | 0.161 | -0.245 | 0.090 | -0.294 | 0.047 |
|---|
| CCNA1 |   | 17 | 30 | 326 | 314 | -0.078 | 0.174 | 0.092 | 0.078 | -0.098 |
|---|
| PPP2R4 |   | 1 | 652 | 1356 | 1357 | 0.174 | -0.011 | -0.013 | 0.054 | -0.129 |
|---|
| H2AFX |   | 4 | 198 | 117 | 125 | -0.086 | 0.111 | -0.072 | 0.121 | -0.010 |
|---|
| CLU |   | 10 | 67 | 117 | 121 | 0.280 | -0.208 | 0.119 | -0.153 | -0.234 |
|---|
| GAK |   | 3 | 265 | 816 | 804 | 0.134 | -0.008 | 0.127 | -0.162 | 0.090 |
|---|
| TGFB1I1 |   | 16 | 34 | 326 | 315 | 0.176 | -0.105 | 0.229 | 0.143 | 0.298 |
|---|
| FGD1 |   | 1 | 652 | 1356 | 1357 | 0.000 | 0.008 | -0.131 | -0.053 | -0.074 |
|---|
| NCOR2 |   | 11 | 59 | 283 | 274 | 0.024 | -0.096 | 0.211 | -0.163 | 0.114 |
|---|
| MRPL44 |   | 1 | 652 | 1356 | 1357 | -0.137 | 0.175 | -0.191 | -0.145 | 0.037 |
|---|
| S100A2 |   | 1 | 652 | 1356 | 1357 | 0.038 | 0.005 | 0.047 | 0.137 | -0.008 |
|---|
| RPL5 |   | 4 | 198 | 889 | 873 | -0.145 | -0.000 | 0.128 | 0.220 | -0.110 |
|---|
| IQGAP2 |   | 16 | 34 | 117 | 119 | 0.264 | -0.170 | -0.051 | -0.112 | 0.049 |
|---|
| GDI1 |   | 1 | 652 | 1356 | 1357 | -0.100 | 0.219 | 0.094 | 0.049 | 0.049 |
|---|
| RB1 |   | 26 | 18 | 271 | 263 | 0.200 | 0.013 | -0.051 | 0.057 | -0.082 |
|---|
| RECQL4 |   | 2 | 391 | 283 | 301 | -0.143 | 0.039 | -0.211 | 0.069 | -0.053 |
|---|
| PPIG |   | 3 | 265 | 563 | 556 | 0.107 | -0.116 | 0.024 | 0.221 | 0.101 |
|---|
| PAK2 |   | 20 | 24 | 1 | 11 | -0.019 | 0.014 | 0.161 | 0.081 | 0.062 |
|---|
| LLGL1 |   | 1 | 652 | 1356 | 1357 | -0.082 | -0.040 | -0.163 | -0.141 | -0.018 |
|---|
| MYO6 |   | 3 | 265 | 923 | 909 | 0.197 | -0.011 | 0.139 | -0.128 | -0.031 |
|---|
| SP1 |   | 16 | 34 | 283 | 272 | -0.129 | 0.222 | -0.254 | 0.239 | 0.082 |
|---|
| PRSS23 |   | 2 | 391 | 778 | 789 | -0.023 | 0.133 | 0.056 | 0.060 | 0.092 |
|---|
| TPM2 |   | 1 | 652 | 1356 | 1357 | 0.147 | 0.007 | 0.269 | 0.212 | 0.203 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| SKP2 |   | 14 | 42 | 148 | 146 | -0.228 | 0.092 | -0.043 | 0.237 | -0.009 |
|---|
| DNASE2 |   | 1 | 652 | 1356 | 1357 | -0.118 | 0.041 | -0.078 | -0.283 | -0.101 |
|---|
| RNF8 |   | 2 | 391 | 117 | 135 | -0.115 | 0.216 | 0.004 | 0.114 | 0.221 |
|---|
| ADAM12 |   | 2 | 391 | 832 | 831 | 0.015 | -0.108 | 0.093 | -0.016 | 0.201 |
|---|
| CCNG1 |   | 4 | 198 | 816 | 802 | 0.287 | -0.100 | 0.061 | -0.157 | -0.093 |
|---|
| MPG |   | 1 | 652 | 1356 | 1357 | 0.124 | 0.043 | 0.044 | -0.282 | 0.121 |
|---|
| SUPT5H |   | 9 | 77 | 1 | 18 | -0.150 | -0.085 | 0.007 | -0.298 | -0.068 |
|---|
| KLF6 |   | 2 | 391 | 1084 | 1072 | 0.295 | 0.173 | 0.001 | 0.099 | 0.115 |
|---|
| PRUNE |   | 1 | 652 | 1356 | 1357 | -0.159 | 0.058 | -0.097 | -0.024 | -0.030 |
|---|
| NR2F1 |   | 6 | 120 | 444 | 438 | 0.086 | -0.252 | 0.113 | 0.133 | 0.207 |
|---|
| DOCK7 |   | 1 | 652 | 1356 | 1357 | undef | 0.043 | 0.032 | undef | undef |
|---|
| PDCD6 |   | 4 | 198 | 444 | 444 | undef | 0.091 | -0.060 | undef | undef |
|---|
| IKBKE |   | 8 | 84 | 756 | 726 | -0.033 | 0.227 | -0.041 | -0.095 | 0.079 |
|---|
| PAX8 |   | 4 | 198 | 778 | 766 | -0.072 | 0.008 | -0.083 | 0.032 | -0.002 |
|---|
| IL7R |   | 14 | 42 | 339 | 334 | 0.130 | -0.151 | -0.112 | -0.130 | -0.160 |
|---|
| S100A1 |   | 3 | 265 | 283 | 292 | -0.045 | 0.244 | 0.044 | 0.013 | 0.063 |
|---|
| HK2 |   | 1 | 652 | 1356 | 1357 | -0.024 | 0.278 | 0.010 | -0.049 | -0.024 |
|---|
| IRF8 |   | 1 | 652 | 1356 | 1357 | 0.141 | -0.007 | -0.193 | -0.050 | -0.067 |
|---|
| ESR1 |   | 4 | 198 | 1292 | 1277 | 0.046 | 0.026 | -0.002 | -0.053 | -0.062 |
|---|
| OSBPL6 |   | 1 | 652 | 1356 | 1357 | undef | 0.028 | 0.135 | undef | undef |
|---|
| CDKN2A |   | 10 | 67 | 364 | 351 | -0.037 | 0.263 | -0.113 | 0.111 | -0.105 |
|---|
| MME |   | 8 | 84 | 57 | 54 | 0.200 | -0.121 | 0.020 | 0.088 | -0.130 |
|---|
GO Enrichment output for subnetwork 941 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.038E-07 | 4.687E-04 |
|---|
| actin filament-based movement | GO:0030048 |  | 7.151E-07 | 8.224E-04 |
|---|
| myeloid cell differentiation | GO:0030099 |  | 8.716E-07 | 6.682E-04 |
|---|
| syncytium formation by plasma membrane fusion | GO:0000768 |  | 1.198E-06 | 6.886E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 1.869E-06 | 8.598E-04 |
|---|
| syncytium formation | GO:0006949 |  | 1.869E-06 | 7.165E-04 |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 2.784E-06 | 9.148E-04 |
|---|
| microspike assembly | GO:0030035 |  | 2.784E-06 | 8.005E-04 |
|---|
| myotube differentiation | GO:0014902 |  | 5.553E-06 | 1.419E-03 |
|---|
| nuclear migration | GO:0007097 |  | 6.748E-06 | 1.552E-03 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 6.748E-06 | 1.411E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 9.984E-08 | 2.439E-04 |
|---|
| actin filament-based movement | GO:0030048 |  | 1.666E-07 | 2.035E-04 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 2.024E-07 | 1.648E-04 |
|---|
| myeloid cell differentiation | GO:0030099 |  | 2.07E-07 | 1.264E-04 |
|---|
| syncytium formation by plasma membrane fusion | GO:0000768 |  | 2.966E-07 | 1.449E-04 |
|---|
| syncytium formation | GO:0006949 |  | 4.638E-07 | 1.889E-04 |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 9.952E-07 | 3.473E-04 |
|---|
| myotube differentiation | GO:0014902 |  | 1.386E-06 | 4.234E-04 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 2.35E-06 | 6.378E-04 |
|---|
| regulation of keratinocyte differentiation | GO:0045616 |  | 2.35E-06 | 5.74E-04 |
|---|
| leukocyte differentiation | GO:0002521 |  | 2.797E-06 | 6.212E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 6.904E-08 | 1.661E-04 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 1.701E-07 | 2.046E-04 |
|---|
| actin filament-based movement | GO:0030048 |  | 1.891E-07 | 1.517E-04 |
|---|
| myeloid cell differentiation | GO:0030099 |  | 2.85E-07 | 1.714E-04 |
|---|
| syncytium formation by plasma membrane fusion | GO:0000768 |  | 4.077E-07 | 1.962E-04 |
|---|
| syncytium formation | GO:0006949 |  | 6.373E-07 | 2.555E-04 |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 1.366E-06 | 4.696E-04 |
|---|
| myotube differentiation | GO:0014902 |  | 1.903E-06 | 5.722E-04 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 2.988E-06 | 7.987E-04 |
|---|
| regulation of keratinocyte differentiation | GO:0045616 |  | 2.988E-06 | 7.188E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 3.423E-06 | 7.487E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.038E-07 | 4.687E-04 |
|---|
| actin filament-based movement | GO:0030048 |  | 7.151E-07 | 8.224E-04 |
|---|
| myeloid cell differentiation | GO:0030099 |  | 8.716E-07 | 6.682E-04 |
|---|
| syncytium formation by plasma membrane fusion | GO:0000768 |  | 1.198E-06 | 6.886E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 1.869E-06 | 8.598E-04 |
|---|
| syncytium formation | GO:0006949 |  | 1.869E-06 | 7.165E-04 |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 2.784E-06 | 9.148E-04 |
|---|
| microspike assembly | GO:0030035 |  | 2.784E-06 | 8.005E-04 |
|---|
| myotube differentiation | GO:0014902 |  | 5.553E-06 | 1.419E-03 |
|---|
| nuclear migration | GO:0007097 |  | 6.748E-06 | 1.552E-03 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 6.748E-06 | 1.411E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.038E-07 | 4.687E-04 |
|---|
| actin filament-based movement | GO:0030048 |  | 7.151E-07 | 8.224E-04 |
|---|
| myeloid cell differentiation | GO:0030099 |  | 8.716E-07 | 6.682E-04 |
|---|
| syncytium formation by plasma membrane fusion | GO:0000768 |  | 1.198E-06 | 6.886E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 1.869E-06 | 8.598E-04 |
|---|
| syncytium formation | GO:0006949 |  | 1.869E-06 | 7.165E-04 |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 2.784E-06 | 9.148E-04 |
|---|
| microspike assembly | GO:0030035 |  | 2.784E-06 | 8.005E-04 |
|---|
| myotube differentiation | GO:0014902 |  | 5.553E-06 | 1.419E-03 |
|---|
| nuclear migration | GO:0007097 |  | 6.748E-06 | 1.552E-03 |
|---|
| regulation of retinoic acid receptor signaling pathway | GO:0048385 |  | 6.748E-06 | 1.411E-03 |
|---|


