Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 639
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.2923 | 8.160e-03 | 4.290e-04 | 3.230e-01 | 1.131e-06 |
|---|
| IPC | 0.4109 | 2.359e-01 | 9.191e-02 | 9.110e-01 | 1.975e-02 |
|---|
| Loi | 0.4387 | 2.930e-04 | 0.000e+00 | 7.508e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6255 | 0.000e+00 | 0.000e+00 | 6.879e-02 | 0.000e+00 |
|---|
| Wang | 0.2389 | 1.545e-02 | 1.026e-01 | 3.153e-01 | 4.998e-04 |
|---|
Expression data for subnetwork 639 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
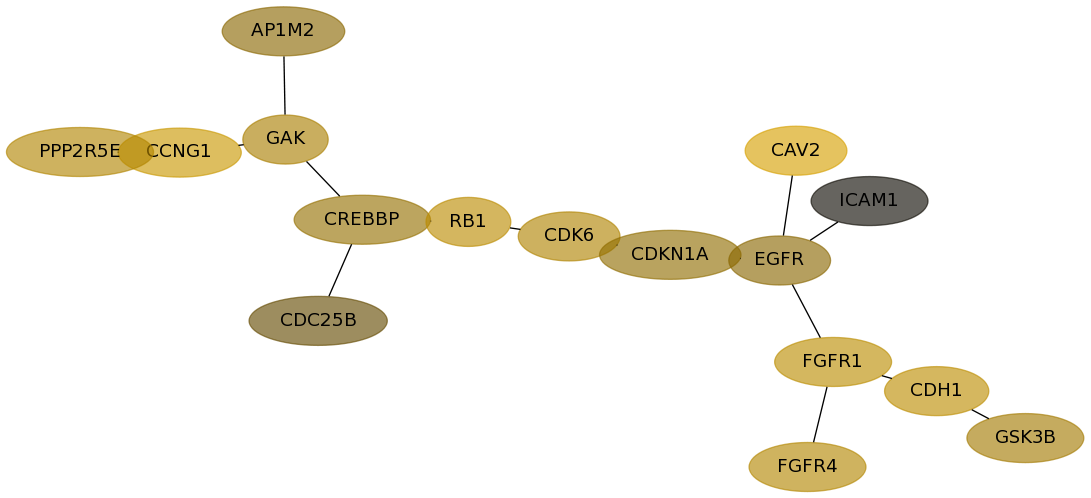
Score for each gene in subnetwork 639 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| CCNG1 |   | 4 | 198 | 816 | 802 | 0.287 | -0.100 | 0.061 | -0.157 | -0.093 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| ICAM1 |   | 2 | 391 | 262 | 283 | 0.001 | 0.147 | -0.054 | -0.040 | -0.132 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| CREBBP |   | 5 | 148 | 444 | 440 | 0.084 | -0.104 | 0.102 | 0.093 | 0.002 |
|---|
| PPP2R5E |   | 3 | 265 | 816 | 804 | 0.160 | 0.001 | 0.191 | 0.018 | 0.094 |
|---|
| RB1 |   | 26 | 18 | 271 | 263 | 0.200 | 0.013 | -0.051 | 0.057 | -0.082 |
|---|
| AP1M2 |   | 2 | 391 | 816 | 808 | 0.064 | -0.082 | 0.154 | -0.068 | 0.056 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| CDC25B |   | 2 | 391 | 206 | 220 | 0.025 | -0.054 | -0.075 | -0.037 | 0.009 |
|---|
| GAK |   | 3 | 265 | 816 | 804 | 0.134 | -0.008 | 0.127 | -0.162 | 0.090 |
|---|
| CDK6 |   | 25 | 19 | 1 | 10 | 0.156 | -0.229 | 0.107 | -0.028 | 0.104 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
GO Enrichment output for subnetwork 639 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.454E-07 | 1.254E-03 |
|---|
| interphase | GO:0051325 |  | 6.119E-07 | 7.037E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.233E-06 | 9.449E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.469E-06 | 1.42E-03 |
|---|
| G1 phase | GO:0051318 |  | 3.375E-06 | 1.552E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 3.872E-06 | 1.484E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 4.093E-06 | 1.345E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 4.154E-06 | 1.194E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 5.089E-06 | 1.3E-03 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 5.817E-06 | 1.338E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 5.817E-06 | 1.216E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.76E-07 | 6.742E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.411E-07 | 6.61E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.295E-06 | 1.055E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 2.799E-06 | 1.71E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 2.936E-06 | 1.434E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.966E-06 | 1.208E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 3.422E-06 | 1.194E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 3.707E-06 | 1.132E-03 |
|---|
| G1 phase | GO:0051318 |  | 3.958E-06 | 1.074E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 3.958E-06 | 9.67E-04 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 5.894E-06 | 1.309E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.474E-07 | 8.359E-04 |
|---|
| interphase | GO:0051325 |  | 4.058E-07 | 4.882E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.414E-07 | 5.946E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.773E-06 | 1.066E-03 |
|---|
| G1 phase | GO:0051318 |  | 3.176E-06 | 1.528E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 3.402E-06 | 1.364E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 3.61E-06 | 1.241E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 3.733E-06 | 1.123E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 3.733E-06 | 9.979E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 4.541E-06 | 1.092E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 5.032E-06 | 1.101E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.454E-07 | 1.254E-03 |
|---|
| interphase | GO:0051325 |  | 6.119E-07 | 7.037E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.233E-06 | 9.449E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.469E-06 | 1.42E-03 |
|---|
| G1 phase | GO:0051318 |  | 3.375E-06 | 1.552E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 3.872E-06 | 1.484E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 4.093E-06 | 1.345E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 4.154E-06 | 1.194E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 5.089E-06 | 1.3E-03 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 5.817E-06 | 1.338E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 5.817E-06 | 1.216E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.454E-07 | 1.254E-03 |
|---|
| interphase | GO:0051325 |  | 6.119E-07 | 7.037E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.233E-06 | 9.449E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.469E-06 | 1.42E-03 |
|---|
| G1 phase | GO:0051318 |  | 3.375E-06 | 1.552E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 3.872E-06 | 1.484E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 4.093E-06 | 1.345E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 4.154E-06 | 1.194E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 5.089E-06 | 1.3E-03 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 5.817E-06 | 1.338E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 5.817E-06 | 1.216E-03 |
|---|


