Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 605
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.2823 | 1.169e-02 | 7.800e-04 | 3.687e-01 | 3.362e-06 |
|---|
| IPC | 0.4279 | 2.106e-01 | 7.598e-02 | 9.050e-01 | 1.448e-02 |
|---|
| Loi | 0.4383 | 2.980e-04 | 0.000e+00 | 7.536e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6328 | 0.000e+00 | 0.000e+00 | 5.893e-02 | 0.000e+00 |
|---|
| Wang | 0.2399 | 1.490e-02 | 1.004e-01 | 3.103e-01 | 4.641e-04 |
|---|
Expression data for subnetwork 605 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
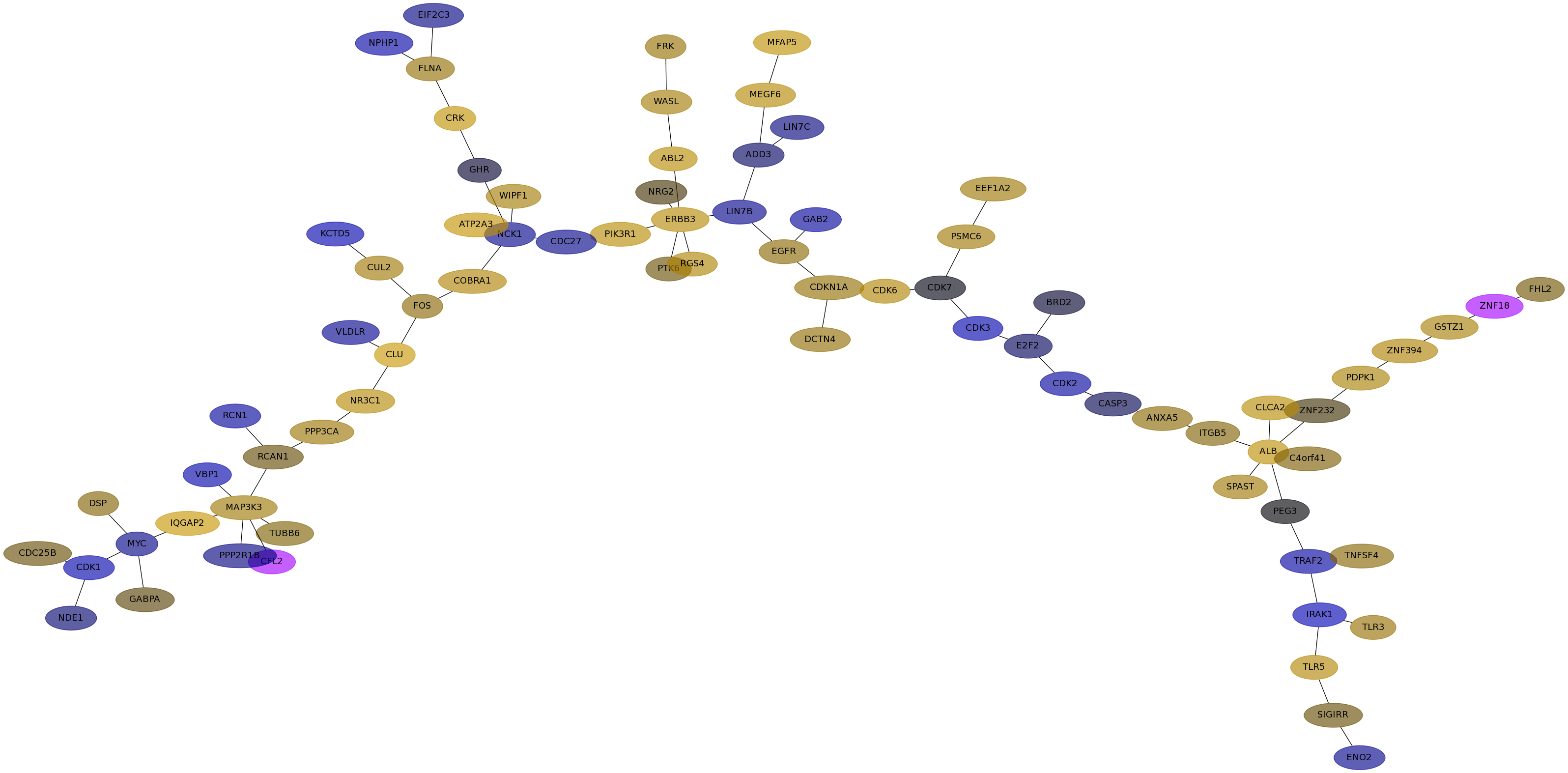
Score for each gene in subnetwork 605 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| PTK6 |   | 2 | 391 | 206 | 220 | 0.029 | -0.010 | 0.204 | -0.343 | 0.213 |
|---|
| RGS4 |   | 3 | 265 | 57 | 71 | 0.144 | 0.022 | 0.160 | -0.006 | 0.371 |
|---|
| SIGIRR |   | 1 | 652 | 206 | 230 | 0.027 | 0.155 | 0.005 | 0.057 | -0.004 |
|---|
| CASP3 |   | 4 | 198 | 129 | 130 | -0.015 | 0.234 | -0.032 | -0.142 | -0.084 |
|---|
| C4orf41 |   | 1 | 652 | 206 | 230 | 0.045 | 0.129 | 0.034 | 0.062 | 0.053 |
|---|
| MFAP5 |   | 1 | 652 | 206 | 230 | 0.222 | 0.195 | 0.036 | 0.040 | 0.142 |
|---|
| NRG2 |   | 1 | 652 | 206 | 230 | 0.011 | -0.088 | 0.050 | -0.095 | -0.005 |
|---|
| TUBB6 |   | 1 | 652 | 206 | 230 | 0.048 | 0.045 | 0.114 | 0.187 | 0.165 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| KCTD5 |   | 1 | 652 | 206 | 230 | -0.143 | -0.038 | -0.128 | -0.143 | 0.060 |
|---|
| CDK3 |   | 5 | 148 | 129 | 129 | -0.151 | -0.076 | 0.189 | -0.041 | 0.082 |
|---|
| PPP2R1B |   | 1 | 652 | 206 | 230 | -0.049 | 0.042 | 0.002 | -0.002 | 0.112 |
|---|
| RCN1 |   | 1 | 652 | 206 | 230 | -0.096 | 0.076 | 0.109 | -0.012 | -0.042 |
|---|
| ABL2 |   | 1 | 652 | 206 | 230 | 0.193 | 0.033 | 0.068 | -0.135 | 0.018 |
|---|
| TLR3 |   | 2 | 391 | 206 | 220 | 0.081 | 0.196 | -0.111 | -0.089 | -0.103 |
|---|
| LIN7B |   | 2 | 391 | 206 | 220 | -0.065 | 0.047 | -0.123 | -0.141 | -0.147 |
|---|
| CDC25B |   | 2 | 391 | 206 | 220 | 0.025 | -0.054 | -0.075 | -0.037 | 0.009 |
|---|
| TLR5 |   | 2 | 391 | 206 | 220 | 0.165 | -0.056 | 0.111 | -0.176 | 0.144 |
|---|
| CDK6 |   | 25 | 19 | 1 | 10 | 0.156 | -0.229 | 0.107 | -0.028 | 0.104 |
|---|
| COBRA1 |   | 4 | 198 | 206 | 205 | 0.150 | 0.137 | 0.335 | 0.159 | -0.016 |
|---|
| MEGF6 |   | 2 | 391 | 206 | 220 | 0.170 | 0.110 | 0.197 | 0.067 | 0.083 |
|---|
| GAB2 |   | 1 | 652 | 206 | 230 | -0.098 | -0.065 | -0.159 | 0.136 | 0.211 |
|---|
| MAP3K3 |   | 4 | 198 | 206 | 205 | 0.102 | 0.085 | 0.046 | 0.028 | 0.094 |
|---|
| ZNF394 |   | 1 | 652 | 206 | 230 | 0.142 | 0.135 | -0.008 | 0.013 | -0.093 |
|---|
| GSTZ1 |   | 1 | 652 | 206 | 230 | 0.126 | 0.107 | -0.129 | -0.016 | -0.080 |
|---|
| GABPA |   | 6 | 120 | 206 | 200 | 0.019 | 0.033 | -0.183 | 0.066 | -0.161 |
|---|
| NCK1 |   | 3 | 265 | 206 | 208 | -0.065 | 0.135 | -0.100 | -0.059 | 0.151 |
|---|
| VBP1 |   | 1 | 652 | 206 | 230 | -0.137 | 0.176 | -0.075 | 0.148 | 0.059 |
|---|
| WIPF1 |   | 1 | 652 | 206 | 230 | 0.112 | 0.020 | -0.108 | -0.043 | 0.114 |
|---|
| FLNA |   | 7 | 100 | 206 | 198 | 0.077 | 0.081 | 0.107 | 0.148 | 0.126 |
|---|
| CDK7 |   | 4 | 198 | 129 | 130 | -0.002 | 0.105 | 0.184 | 0.017 | -0.097 |
|---|
| PEG3 |   | 1 | 652 | 206 | 230 | -0.001 | 0.034 | 0.099 | 0.065 | 0.030 |
|---|
| FRK |   | 1 | 652 | 206 | 230 | 0.082 | 0.110 | 0.003 | -0.156 | 0.190 |
|---|
| DSP |   | 7 | 100 | 206 | 198 | 0.052 | 0.054 | 0.235 | -0.119 | 0.053 |
|---|
| CDK1 |   | 16 | 34 | 206 | 194 | -0.138 | 0.265 | -0.195 | 0.106 | 0.031 |
|---|
| ZNF232 |   | 5 | 148 | 57 | 63 | 0.010 | 0.048 | -0.040 | 0.189 | 0.159 |
|---|
| CRK |   | 27 | 17 | 206 | 193 | 0.232 | -0.116 | 0.106 | 0.090 | 0.017 |
|---|
| BRD2 |   | 3 | 265 | 129 | 139 | -0.006 | 0.016 | 0.348 | 0.105 | 0.093 |
|---|
| PSMC6 |   | 1 | 652 | 206 | 230 | 0.106 | 0.163 | 0.061 | -0.010 | 0.044 |
|---|
| WASL |   | 2 | 391 | 206 | 220 | 0.111 | 0.186 | -0.065 | -0.021 | -0.084 |
|---|
| CLCA2 |   | 5 | 148 | 206 | 202 | 0.188 | -0.176 | 0.243 | -0.153 | -0.000 |
|---|
| ZNF18 |   | 1 | 652 | 206 | 230 | undef | 0.085 | 0.072 | undef | undef |
|---|
| CLU |   | 10 | 67 | 117 | 121 | 0.280 | -0.208 | 0.119 | -0.153 | -0.234 |
|---|
| TNFSF4 |   | 1 | 652 | 206 | 230 | 0.057 | 0.229 | -0.071 | -0.191 | 0.160 |
|---|
| E2F2 |   | 4 | 198 | 129 | 130 | -0.021 | 0.212 | -0.047 | 0.162 | -0.092 |
|---|
| ENO2 |   | 2 | 391 | 206 | 220 | -0.066 | 0.085 | 0.114 | 0.148 | 0.208 |
|---|
| TRAF2 |   | 6 | 120 | 206 | 200 | -0.107 | 0.185 | -0.172 | -0.013 | -0.092 |
|---|
| ATP2A3 |   | 9 | 77 | 1 | 18 | 0.240 | -0.040 | 0.082 | -0.229 | -0.025 |
|---|
| RCAN1 |   | 1 | 652 | 206 | 230 | 0.025 | -0.002 | 0.111 | 0.005 | -0.045 |
|---|
| ITGB5 |   | 1 | 652 | 206 | 230 | 0.051 | 0.163 | 0.208 | 0.081 | 0.148 |
|---|
| IQGAP2 |   | 16 | 34 | 117 | 119 | 0.264 | -0.170 | -0.051 | -0.112 | 0.049 |
|---|
| ADD3 |   | 1 | 652 | 206 | 230 | -0.024 | -0.274 | 0.035 | -0.192 | 0.183 |
|---|
| IRAK1 |   | 4 | 198 | 206 | 205 | -0.170 | 0.266 | -0.023 | 0.092 | -0.102 |
|---|
| MYC |   | 24 | 21 | 148 | 137 | -0.056 | -0.101 | -0.169 | 0.188 | -0.134 |
|---|
| EEF1A2 |   | 9 | 77 | 182 | 179 | 0.098 | 0.032 | 0.220 | -0.119 | 0.102 |
|---|
| GHR |   | 14 | 42 | 148 | 146 | -0.006 | -0.112 | 0.174 | 0.090 | 0.201 |
|---|
| DCTN4 |   | 1 | 652 | 206 | 230 | 0.073 | 0.122 | -0.025 | -0.111 | -0.105 |
|---|
| ANXA5 |   | 1 | 652 | 206 | 230 | 0.064 | 0.098 | -0.009 | 0.126 | 0.264 |
|---|
| ERBB3 |   | 5 | 148 | 206 | 202 | 0.175 | -0.171 | -0.007 | -0.141 | -0.098 |
|---|
| PDPK1 |   | 1 | 652 | 206 | 230 | 0.130 | -0.050 | -0.200 | -0.024 | 0.134 |
|---|
| PPP3CA |   | 1 | 652 | 206 | 230 | 0.094 | 0.015 | 0.140 | 0.110 | 0.042 |
|---|
| NR3C1 |   | 1 | 652 | 206 | 230 | 0.174 | 0.026 | -0.019 | -0.047 | 0.197 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| VLDLR |   | 1 | 652 | 206 | 230 | -0.067 | -0.059 | 0.109 | -0.128 | 0.024 |
|---|
| FHL2 |   | 5 | 148 | 206 | 202 | 0.033 | 0.143 | 0.239 | -0.033 | 0.294 |
|---|
| FOS |   | 20 | 24 | 148 | 138 | 0.060 | -0.137 | 0.129 | 0.185 | 0.234 |
|---|
| LIN7C |   | 1 | 652 | 206 | 230 | -0.043 | 0.166 | 0.088 | 0.118 | 0.094 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| CFL2 |   | 1 | 652 | 206 | 230 | undef | 0.235 | 0.026 | undef | undef |
|---|
| CDC27 |   | 4 | 198 | 1 | 23 | -0.062 | 0.156 | -0.086 | -0.029 | 0.348 |
|---|
| ALB |   | 7 | 100 | 57 | 60 | 0.207 | -0.122 | 0.218 | 0.080 | 0.038 |
|---|
| CUL2 |   | 3 | 265 | 206 | 208 | 0.103 | 0.124 | -0.192 | -0.090 | 0.166 |
|---|
| NDE1 |   | 1 | 652 | 206 | 230 | -0.032 | 0.142 | -0.058 | 0.142 | 0.111 |
|---|
| CDK2 |   | 16 | 34 | 129 | 124 | -0.104 | 0.196 | -0.096 | 0.109 | 0.362 |
|---|
| NPHP1 |   | 2 | 391 | 206 | 220 | -0.113 | 0.139 | 0.095 | 0.026 | -0.110 |
|---|
| EIF2C3 |   | 2 | 391 | 206 | 220 | -0.053 | -0.198 | 0.070 | 0.040 | -0.125 |
|---|
| SPAST |   | 1 | 652 | 206 | 230 | 0.104 | 0.033 | 0.029 | -0.271 | 0.135 |
|---|
GO Enrichment output for subnetwork 605 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.051E-07 | 4.718E-04 |
|---|
| interphase | GO:0051325 |  | 2.455E-07 | 2.823E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.385E-07 | 4.895E-04 |
|---|
| activation of NF-kappaB-inducing kinase activity | GO:0007250 |  | 2.513E-06 | 1.445E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.003E-05 | 4.615E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.042E-05 | 7.827E-03 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.042E-05 | 6.709E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.36E-05 | 9.661E-03 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 4.329E-05 | 0.01106239 |
|---|
| regulation of chemokine biosynthetic process | GO:0045073 |  | 4.329E-05 | 9.956E-03 |
|---|
| actin filament organization | GO:0007015 |  | 6.837E-05 | 0.01429506 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.333E-08 | 1.791E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.97E-07 | 2.406E-04 |
|---|
| activation of NF-kappaB-inducing kinase activity | GO:0007250 |  | 1.977E-06 | 1.61E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.826E-06 | 2.948E-03 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.333E-05 | 6.513E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.396E-05 | 5.686E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.568E-05 | 5.473E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 1.897E-05 | 5.792E-03 |
|---|
| regulation of chemokine biosynthetic process | GO:0045073 |  | 1.897E-05 | 5.148E-03 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 3.449E-05 | 8.426E-03 |
|---|
| regulation of chemokine production | GO:0032642 |  | 4.466E-05 | 9.918E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.184E-07 | 2.848E-04 |
|---|
| interphase | GO:0051325 |  | 1.509E-07 | 1.815E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.436E-07 | 2.756E-04 |
|---|
| activation of NF-kappaB-inducing kinase activity | GO:0007250 |  | 2.888E-06 | 1.737E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 7.66E-06 | 3.686E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.738E-05 | 6.971E-03 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.775E-05 | 6.102E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.476E-05 | 7.446E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.525E-05 | 6.75E-03 |
|---|
| regulation of chemokine biosynthetic process | GO:0045073 |  | 2.525E-05 | 6.075E-03 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 4.588E-05 | 0.01003486 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.051E-07 | 4.718E-04 |
|---|
| interphase | GO:0051325 |  | 2.455E-07 | 2.823E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.385E-07 | 4.895E-04 |
|---|
| activation of NF-kappaB-inducing kinase activity | GO:0007250 |  | 2.513E-06 | 1.445E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.003E-05 | 4.615E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.042E-05 | 7.827E-03 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.042E-05 | 6.709E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.36E-05 | 9.661E-03 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 4.329E-05 | 0.01106239 |
|---|
| regulation of chemokine biosynthetic process | GO:0045073 |  | 4.329E-05 | 9.956E-03 |
|---|
| actin filament organization | GO:0007015 |  | 6.837E-05 | 0.01429506 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.051E-07 | 4.718E-04 |
|---|
| interphase | GO:0051325 |  | 2.455E-07 | 2.823E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.385E-07 | 4.895E-04 |
|---|
| activation of NF-kappaB-inducing kinase activity | GO:0007250 |  | 2.513E-06 | 1.445E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.003E-05 | 4.615E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.042E-05 | 7.827E-03 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.042E-05 | 6.709E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.36E-05 | 9.661E-03 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 4.329E-05 | 0.01106239 |
|---|
| regulation of chemokine biosynthetic process | GO:0045073 |  | 4.329E-05 | 9.956E-03 |
|---|
| actin filament organization | GO:0007015 |  | 6.837E-05 | 0.01429506 |
|---|


