Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 573
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.2761 | 1.450e-02 | 1.116e-03 | 3.979e-01 | 6.438e-06 |
|---|
| IPC | 0.4426 | 1.897e-01 | 6.375e-02 | 8.999e-01 | 1.088e-02 |
|---|
| Loi | 0.4334 | 3.650e-04 | 0.000e+00 | 7.861e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6360 | 0.000e+00 | 0.000e+00 | 5.495e-02 | 0.000e+00 |
|---|
| Wang | 0.2365 | 1.683e-02 | 1.082e-01 | 3.272e-01 | 5.959e-04 |
|---|
Expression data for subnetwork 573 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
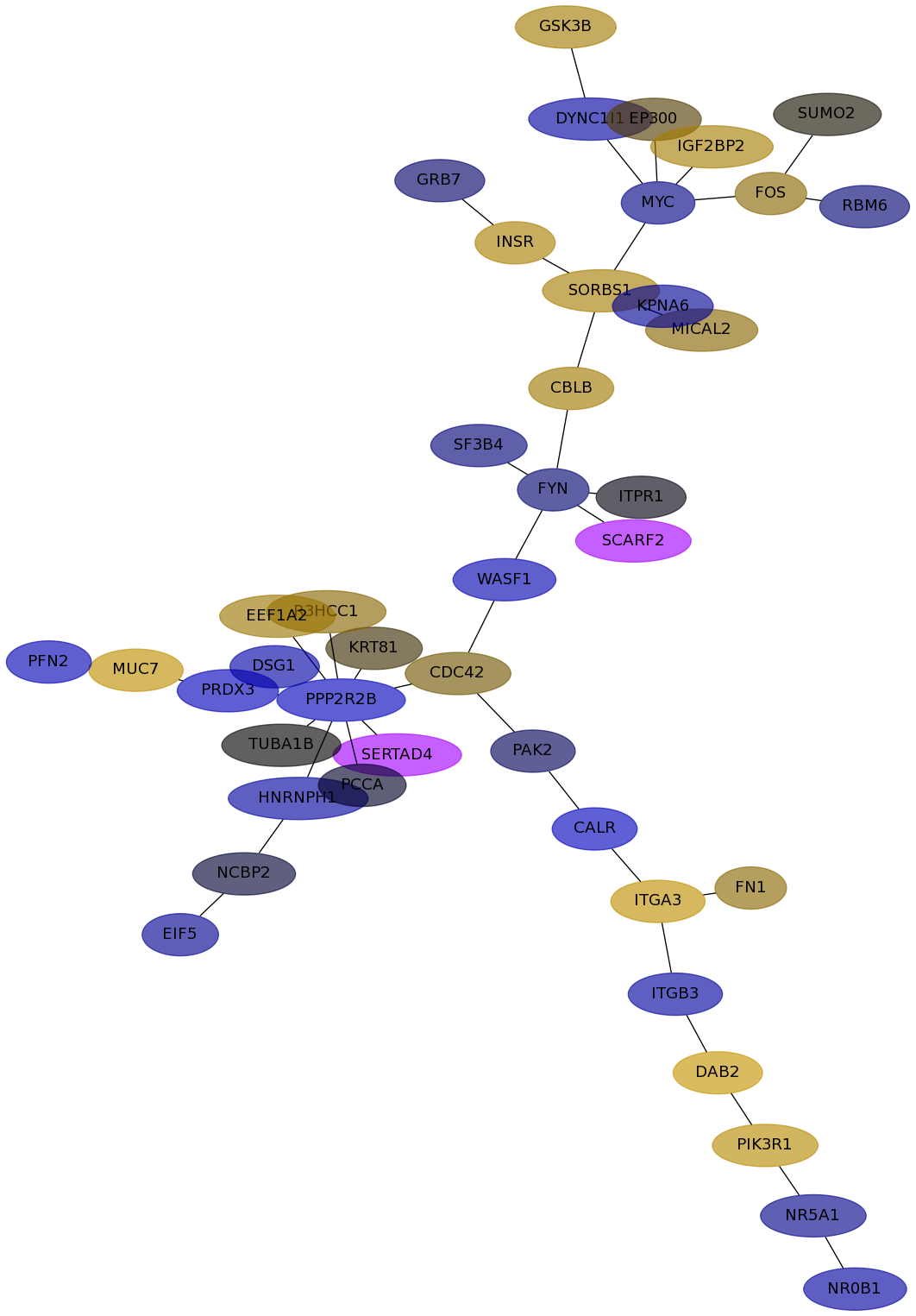
Score for each gene in subnetwork 573 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| SORBS1 |   | 17 | 30 | 182 | 178 | 0.124 | -0.043 | 0.015 | 0.314 | 0.200 |
|---|
| FN1 |   | 5 | 148 | 57 | 63 | 0.064 | 0.190 | 0.133 | 0.091 | 0.083 |
|---|
| WASF1 |   | 2 | 391 | 182 | 195 | -0.159 | 0.162 | -0.070 | 0.348 | 0.130 |
|---|
| SUMO2 |   | 12 | 55 | 148 | 150 | 0.003 | 0.117 | 0.062 | 0.294 | -0.096 |
|---|
| IGF2BP2 |   | 3 | 265 | 148 | 166 | 0.124 | 0.210 | -0.002 | 0.098 | -0.122 |
|---|
| MUC7 |   | 1 | 652 | 182 | 210 | 0.219 | -0.113 | 0.056 | 0.100 | -0.175 |
|---|
| HNRNPH1 |   | 1 | 652 | 182 | 210 | -0.100 | -0.059 | 0.248 | 0.125 | -0.133 |
|---|
| PFN2 |   | 4 | 198 | 182 | 183 | -0.164 | 0.014 | -0.001 | 0.286 | -0.056 |
|---|
| KRT81 |   | 2 | 391 | 182 | 195 | 0.009 | 0.108 | -0.105 | 0.252 | 0.086 |
|---|
| DYNC1I1 |   | 5 | 148 | 182 | 181 | -0.109 | -0.069 | 0.013 | -0.086 | 0.088 |
|---|
| CALR |   | 10 | 67 | 117 | 121 | -0.216 | -0.105 | 0.002 | -0.061 | -0.075 |
|---|
| NR0B1 |   | 1 | 652 | 182 | 210 | -0.106 | 0.135 | -0.035 | 0.000 | 0.153 |
|---|
| DAB2 |   | 13 | 47 | 1 | 15 | 0.258 | -0.175 | 0.067 | -0.022 | 0.071 |
|---|
| ITGA3 |   | 3 | 265 | 1 | 24 | 0.218 | -0.135 | 0.299 | 0.037 | -0.104 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| MYC |   | 24 | 21 | 148 | 137 | -0.056 | -0.101 | -0.169 | 0.188 | -0.134 |
|---|
| EEF1A2 |   | 9 | 77 | 182 | 179 | 0.098 | 0.032 | 0.220 | -0.119 | 0.102 |
|---|
| PAK2 |   | 20 | 24 | 1 | 11 | -0.019 | 0.014 | 0.161 | 0.081 | 0.062 |
|---|
| RBM6 |   | 10 | 67 | 148 | 152 | -0.036 | -0.082 | 0.265 | -0.004 | 0.111 |
|---|
| ITGB3 |   | 3 | 265 | 1 | 24 | -0.100 | -0.015 | 0.030 | 0.086 | 0.085 |
|---|
| NR5A1 |   | 1 | 652 | 182 | 210 | -0.062 | 0.135 | -0.081 | -0.080 | -0.008 |
|---|
| SCARF2 |   | 1 | 652 | 182 | 210 | undef | -0.114 | 0.148 | undef | undef |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| EIF5 |   | 1 | 652 | 182 | 210 | -0.073 | -0.140 | -0.084 | 0.264 | -0.178 |
|---|
| KPNA6 |   | 5 | 148 | 182 | 181 | -0.082 | -0.144 | 0.128 | -0.005 | -0.108 |
|---|
| MICAL2 |   | 3 | 265 | 182 | 189 | 0.061 | -0.012 | 0.143 | -0.042 | 0.161 |
|---|
| PCCA |   | 1 | 652 | 182 | 210 | -0.005 | -0.045 | 0.042 | 0.100 | -0.016 |
|---|
| SERTAD4 |   | 7 | 100 | 1 | 20 | undef | 0.173 | 0.169 | undef | undef |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| CBLB |   | 3 | 265 | 182 | 189 | 0.109 | -0.168 | 0.266 | -0.248 | 0.223 |
|---|
| R3HCC1 |   | 1 | 652 | 182 | 210 | 0.060 | 0.045 | 0.058 | 0.158 | -0.025 |
|---|
| FOS |   | 20 | 24 | 148 | 138 | 0.060 | -0.137 | 0.129 | 0.185 | 0.234 |
|---|
| TUBA1B |   | 7 | 100 | 182 | 180 | 0.001 | 0.116 | 0.094 | 0.249 | 0.128 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| FYN |   | 3 | 265 | 182 | 189 | -0.032 | -0.171 | -0.167 | 0.073 | 0.110 |
|---|
| SF3B4 |   | 1 | 652 | 182 | 210 | -0.042 | -0.017 | 0.017 | -0.199 | -0.007 |
|---|
| PRDX3 |   | 2 | 391 | 182 | 195 | -0.194 | 0.038 | -0.059 | 0.173 | -0.143 |
|---|
| ITPR1 |   | 3 | 265 | 182 | 189 | -0.002 | -0.096 | 0.077 | 0.015 | 0.008 |
|---|
| GRB7 |   | 13 | 47 | 57 | 51 | -0.028 | 0.021 | 0.115 | -0.072 | 0.129 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| NCBP2 |   | 1 | 652 | 182 | 210 | -0.007 | -0.087 | 0.170 | 0.311 | -0.052 |
|---|
| DSG1 |   | 4 | 198 | 182 | 183 | -0.120 | 0.072 | -0.075 | 0.252 | 0.068 |
|---|
GO Enrichment output for subnetwork 573 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 1.592E-06 | 3.662E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.592E-06 | 1.831E-03 |
|---|
| regulation of nucleobase. nucleoside. nucleotide and nucleic acid transport | GO:0032239 |  | 1.592E-06 | 1.221E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 1.924E-06 | 1.106E-03 |
|---|
| sex determination | GO:0007530 |  | 3.107E-06 | 1.429E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 3.107E-06 | 1.191E-03 |
|---|
| male sex determination | GO:0030238 |  | 3.171E-06 | 1.042E-03 |
|---|
| macrophage differentiation | GO:0030225 |  | 3.171E-06 | 9.118E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 5.528E-06 | 1.413E-03 |
|---|
| adrenal gland development | GO:0030325 |  | 5.528E-06 | 1.271E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 5.528E-06 | 1.156E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nucleobase. nucleoside. nucleotide and nucleic acid transport | GO:0032239 |  | 4.327E-07 | 1.057E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 6.979E-07 | 8.525E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 6.979E-07 | 5.683E-04 |
|---|
| nuclear migration | GO:0007097 |  | 8.631E-07 | 5.272E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 8.631E-07 | 4.217E-04 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 1.048E-06 | 4.267E-04 |
|---|
| adrenal gland development | GO:0030325 |  | 1.507E-06 | 5.258E-04 |
|---|
| male sex determination | GO:0030238 |  | 1.507E-06 | 4.601E-04 |
|---|
| sex determination | GO:0007530 |  | 1.514E-06 | 4.11E-04 |
|---|
| interaction with host | GO:0051701 |  | 1.797E-06 | 4.391E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.404E-06 | 5.34E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nucleobase. nucleoside. nucleotide and nucleic acid transport | GO:0032239 |  | 6.289E-07 | 1.513E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 1.143E-06 | 1.375E-03 |
|---|
| negative regulation of translation | GO:0017148 |  | 1.143E-06 | 9.164E-04 |
|---|
| nuclear migration | GO:0007097 |  | 1.254E-06 | 7.543E-04 |
|---|
| male sex determination | GO:0030238 |  | 1.254E-06 | 6.035E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.254E-06 | 5.029E-04 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 1.714E-06 | 5.893E-04 |
|---|
| sex determination | GO:0007530 |  | 2.069E-06 | 6.223E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.188E-06 | 5.85E-04 |
|---|
| adrenal gland development | GO:0030325 |  | 2.188E-06 | 5.265E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 2.188E-06 | 4.786E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 1.592E-06 | 3.662E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.592E-06 | 1.831E-03 |
|---|
| regulation of nucleobase. nucleoside. nucleotide and nucleic acid transport | GO:0032239 |  | 1.592E-06 | 1.221E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 1.924E-06 | 1.106E-03 |
|---|
| sex determination | GO:0007530 |  | 3.107E-06 | 1.429E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 3.107E-06 | 1.191E-03 |
|---|
| male sex determination | GO:0030238 |  | 3.171E-06 | 1.042E-03 |
|---|
| macrophage differentiation | GO:0030225 |  | 3.171E-06 | 9.118E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 5.528E-06 | 1.413E-03 |
|---|
| adrenal gland development | GO:0030325 |  | 5.528E-06 | 1.271E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 5.528E-06 | 1.156E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 1.592E-06 | 3.662E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.592E-06 | 1.831E-03 |
|---|
| regulation of nucleobase. nucleoside. nucleotide and nucleic acid transport | GO:0032239 |  | 1.592E-06 | 1.221E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 1.924E-06 | 1.106E-03 |
|---|
| sex determination | GO:0007530 |  | 3.107E-06 | 1.429E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 3.107E-06 | 1.191E-03 |
|---|
| male sex determination | GO:0030238 |  | 3.171E-06 | 1.042E-03 |
|---|
| macrophage differentiation | GO:0030225 |  | 3.171E-06 | 9.118E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 5.528E-06 | 1.413E-03 |
|---|
| adrenal gland development | GO:0030325 |  | 5.528E-06 | 1.271E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 5.528E-06 | 1.156E-03 |
|---|


