Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 532
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.2811 | 1.221e-02 | 8.380e-04 | 3.745e-01 | 3.832e-06 |
|---|
| IPC | 0.4490 | 1.809e-01 | 5.890e-02 | 8.977e-01 | 9.564e-03 |
|---|
| Loi | 0.4259 | 4.960e-04 | 0.000e+00 | 8.385e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6205 | 0.000e+00 | 0.000e+00 | 7.609e-02 | 0.000e+00 |
|---|
| Wang | 0.2322 | 1.958e-02 | 1.187e-01 | 3.491e-01 | 8.115e-04 |
|---|
Expression data for subnetwork 532 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
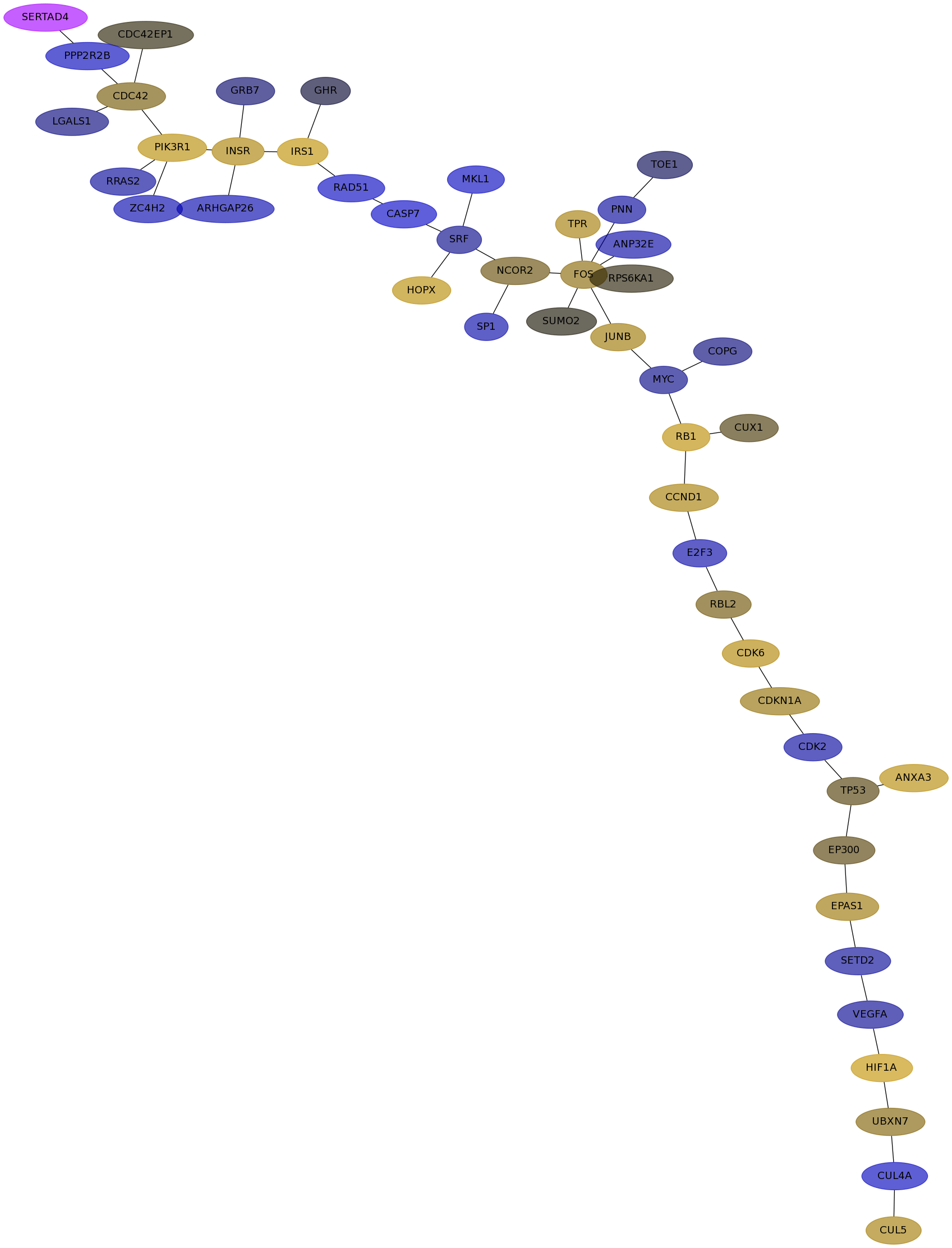
Score for each gene in subnetwork 532 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| ARHGAP26 |   | 5 | 148 | 444 | 440 | -0.145 | -0.023 | 0.171 | -0.222 | -0.348 |
|---|
| ANXA3 |   | 1 | 652 | 696 | 716 | 0.182 | 0.152 | 0.132 | 0.194 | 0.030 |
|---|
| NCOR2 |   | 11 | 59 | 283 | 274 | 0.024 | -0.096 | 0.211 | -0.163 | 0.114 |
|---|
| CUL4A |   | 1 | 652 | 696 | 716 | -0.209 | -0.031 | -0.058 | 0.112 | -0.100 |
|---|
| EPAS1 |   | 1 | 652 | 696 | 716 | 0.093 | -0.144 | 0.073 | -0.023 | 0.141 |
|---|
| RPS6KA1 |   | 8 | 84 | 696 | 669 | 0.005 | -0.208 | 0.084 | -0.322 | -0.071 |
|---|
| SUMO2 |   | 12 | 55 | 148 | 150 | 0.003 | 0.117 | 0.062 | 0.294 | -0.096 |
|---|
| MKL1 |   | 1 | 652 | 696 | 716 | -0.219 | 0.221 | 0.174 | 0.084 | -0.091 |
|---|
| TP53 |   | 28 | 15 | 1 | 9 | 0.015 | -0.167 | 0.148 | -0.027 | 0.147 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| RBL2 |   | 2 | 391 | 492 | 506 | 0.031 | -0.101 | 0.012 | -0.100 | 0.167 |
|---|
| HOPX |   | 1 | 652 | 696 | 716 | 0.190 | 0.137 | 0.121 | -0.128 | 0.234 |
|---|
| UBXN7 |   | 6 | 120 | 148 | 156 | 0.049 | -0.055 | 0.153 | 0.224 | 0.175 |
|---|
| CCND1 |   | 12 | 55 | 148 | 150 | 0.122 | -0.258 | 0.074 | 0.117 | 0.003 |
|---|
| RB1 |   | 26 | 18 | 271 | 263 | 0.200 | 0.013 | -0.051 | 0.057 | -0.082 |
|---|
| CUL5 |   | 4 | 198 | 492 | 499 | 0.111 | 0.051 | 0.187 | 0.221 | 0.216 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| MYC |   | 24 | 21 | 148 | 137 | -0.056 | -0.101 | -0.169 | 0.188 | -0.134 |
|---|
| JUNB |   | 5 | 148 | 696 | 672 | 0.101 | 0.057 | 0.086 | 0.065 | -0.015 |
|---|
| GHR |   | 14 | 42 | 148 | 146 | -0.006 | -0.112 | 0.174 | 0.090 | 0.201 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| RRAS2 |   | 6 | 120 | 563 | 549 | -0.089 | 0.155 | 0.076 | 0.289 | 0.031 |
|---|
| CDK6 |   | 25 | 19 | 1 | 10 | 0.156 | -0.229 | 0.107 | -0.028 | 0.104 |
|---|
| SP1 |   | 16 | 34 | 283 | 272 | -0.129 | 0.222 | -0.254 | 0.239 | 0.082 |
|---|
| ZC4H2 |   | 16 | 34 | 339 | 332 | -0.145 | 0.065 | 0.061 | 0.304 | 0.059 |
|---|
| SERTAD4 |   | 7 | 100 | 1 | 20 | undef | 0.173 | 0.169 | undef | undef |
|---|
| SETD2 |   | 2 | 391 | 696 | 698 | -0.081 | -0.129 | 0.075 | 0.038 | 0.138 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| CUX1 |   | 8 | 84 | 271 | 266 | 0.012 | -0.042 | 0.256 | -0.121 | 0.040 |
|---|
| E2F3 |   | 5 | 148 | 364 | 358 | -0.126 | 0.041 | -0.091 | 0.177 | 0.050 |
|---|
| LGALS1 |   | 3 | 265 | 696 | 686 | -0.045 | 0.267 | 0.010 | 0.218 | 0.156 |
|---|
| PNN |   | 1 | 652 | 696 | 716 | -0.092 | 0.051 | 0.175 | 0.218 | -0.186 |
|---|
| VEGFA |   | 28 | 15 | 57 | 49 | -0.074 | 0.090 | 0.200 | 0.030 | 0.148 |
|---|
| FOS |   | 20 | 24 | 148 | 138 | 0.060 | -0.137 | 0.129 | 0.185 | 0.234 |
|---|
| IRS1 |   | 14 | 42 | 1 | 14 | 0.217 | -0.219 | 0.065 | 0.165 | 0.064 |
|---|
| TOE1 |   | 1 | 652 | 696 | 716 | -0.016 | -0.049 | -0.185 | 0.162 | -0.052 |
|---|
| TPR |   | 10 | 67 | 148 | 152 | 0.118 | -0.130 | 0.120 | -0.036 | 0.156 |
|---|
| CASP7 |   | 8 | 84 | 283 | 278 | -0.270 | 0.216 | 0.095 | 0.255 | 0.035 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| HIF1A |   | 21 | 23 | 33 | 31 | 0.246 | -0.098 | 0.224 | -0.118 | 0.356 |
|---|
| RAD51 |   | 4 | 198 | 696 | 684 | -0.225 | 0.146 | -0.098 | 0.135 | 0.061 |
|---|
| GRB7 |   | 13 | 47 | 57 | 51 | -0.028 | 0.021 | 0.115 | -0.072 | 0.129 |
|---|
| SRF |   | 3 | 265 | 283 | 292 | -0.061 | -0.017 | -0.023 | 0.309 | 0.108 |
|---|
| CDC42EP1 |   | 1 | 652 | 696 | 716 | 0.005 | 0.138 | 0.049 | 0.065 | 0.050 |
|---|
| ANP32E |   | 6 | 120 | 148 | 156 | -0.114 | 0.166 | -0.139 | 0.191 | 0.118 |
|---|
| CDK2 |   | 16 | 34 | 129 | 124 | -0.104 | 0.196 | -0.096 | 0.109 | 0.362 |
|---|
| COPG |   | 2 | 391 | 696 | 698 | -0.040 | 0.004 | 0.080 | -0.158 | 0.280 |
|---|
GO Enrichment output for subnetwork 532 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 7.698E-14 | 1.771E-10 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 2.593E-09 | 2.981E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 8.703E-09 | 6.672E-06 |
|---|
| interphase | GO:0051325 |  | 1.046E-08 | 6.015E-06 |
|---|
| trophectodermal cell differentiation | GO:0001829 |  | 1.801E-08 | 8.286E-06 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 3.589E-08 | 1.376E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.714E-08 | 1.22E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.831E-08 | 1.676E-05 |
|---|
| blastocyst formation | GO:0001825 |  | 6.437E-08 | 1.645E-05 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 1.069E-07 | 2.458E-05 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.67E-07 | 3.491E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 5.929E-14 | 1.448E-10 |
|---|
| lung development | GO:0030324 |  | 1.785E-10 | 2.181E-07 |
|---|
| respiratory tube development | GO:0030323 |  | 2.532E-10 | 2.062E-07 |
|---|
| respiratory system development | GO:0060541 |  | 3.169E-10 | 1.936E-07 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 2.548E-09 | 1.245E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.075E-09 | 1.252E-06 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 3.618E-09 | 1.263E-06 |
|---|
| trophectodermal cell differentiation | GO:0001829 |  | 1.182E-08 | 3.61E-06 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 1.517E-08 | 4.119E-06 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 1.517E-08 | 3.707E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.544E-08 | 3.429E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.22E-14 | 5.34E-11 |
|---|
| lung development | GO:0030324 |  | 2.348E-10 | 2.825E-07 |
|---|
| respiratory tube development | GO:0030323 |  | 3.366E-10 | 2.699E-07 |
|---|
| respiratory system development | GO:0060541 |  | 4.24E-10 | 2.55E-07 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.338E-09 | 1.606E-06 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 3.577E-09 | 1.434E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.996E-09 | 1.373E-06 |
|---|
| interphase | GO:0051325 |  | 5.123E-09 | 1.541E-06 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 1.254E-08 | 3.352E-06 |
|---|
| trophectodermal cell differentiation | GO:0001829 |  | 1.548E-08 | 3.726E-06 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 1.641E-08 | 3.588E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 7.698E-14 | 1.771E-10 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 2.593E-09 | 2.981E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 8.703E-09 | 6.672E-06 |
|---|
| interphase | GO:0051325 |  | 1.046E-08 | 6.015E-06 |
|---|
| trophectodermal cell differentiation | GO:0001829 |  | 1.801E-08 | 8.286E-06 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 3.589E-08 | 1.376E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.714E-08 | 1.22E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.831E-08 | 1.676E-05 |
|---|
| blastocyst formation | GO:0001825 |  | 6.437E-08 | 1.645E-05 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 1.069E-07 | 2.458E-05 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.67E-07 | 3.491E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 7.698E-14 | 1.771E-10 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 2.593E-09 | 2.981E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 8.703E-09 | 6.672E-06 |
|---|
| interphase | GO:0051325 |  | 1.046E-08 | 6.015E-06 |
|---|
| trophectodermal cell differentiation | GO:0001829 |  | 1.801E-08 | 8.286E-06 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 3.589E-08 | 1.376E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.714E-08 | 1.22E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.831E-08 | 1.676E-05 |
|---|
| blastocyst formation | GO:0001825 |  | 6.437E-08 | 1.645E-05 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 1.069E-07 | 2.458E-05 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.67E-07 | 3.491E-05 |
|---|


