Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 528
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.2806 | 1.241e-02 | 8.610e-04 | 3.767e-01 | 4.025e-06 |
|---|
| IPC | 0.4493 | 1.805e-01 | 5.868e-02 | 8.976e-01 | 9.506e-03 |
|---|
| Loi | 0.4250 | 5.140e-04 | 0.000e+00 | 8.449e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6189 | 0.000e+00 | 0.000e+00 | 7.856e-02 | 0.000e+00 |
|---|
| Wang | 0.2319 | 1.983e-02 | 1.197e-01 | 3.510e-01 | 8.328e-04 |
|---|
Expression data for subnetwork 528 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
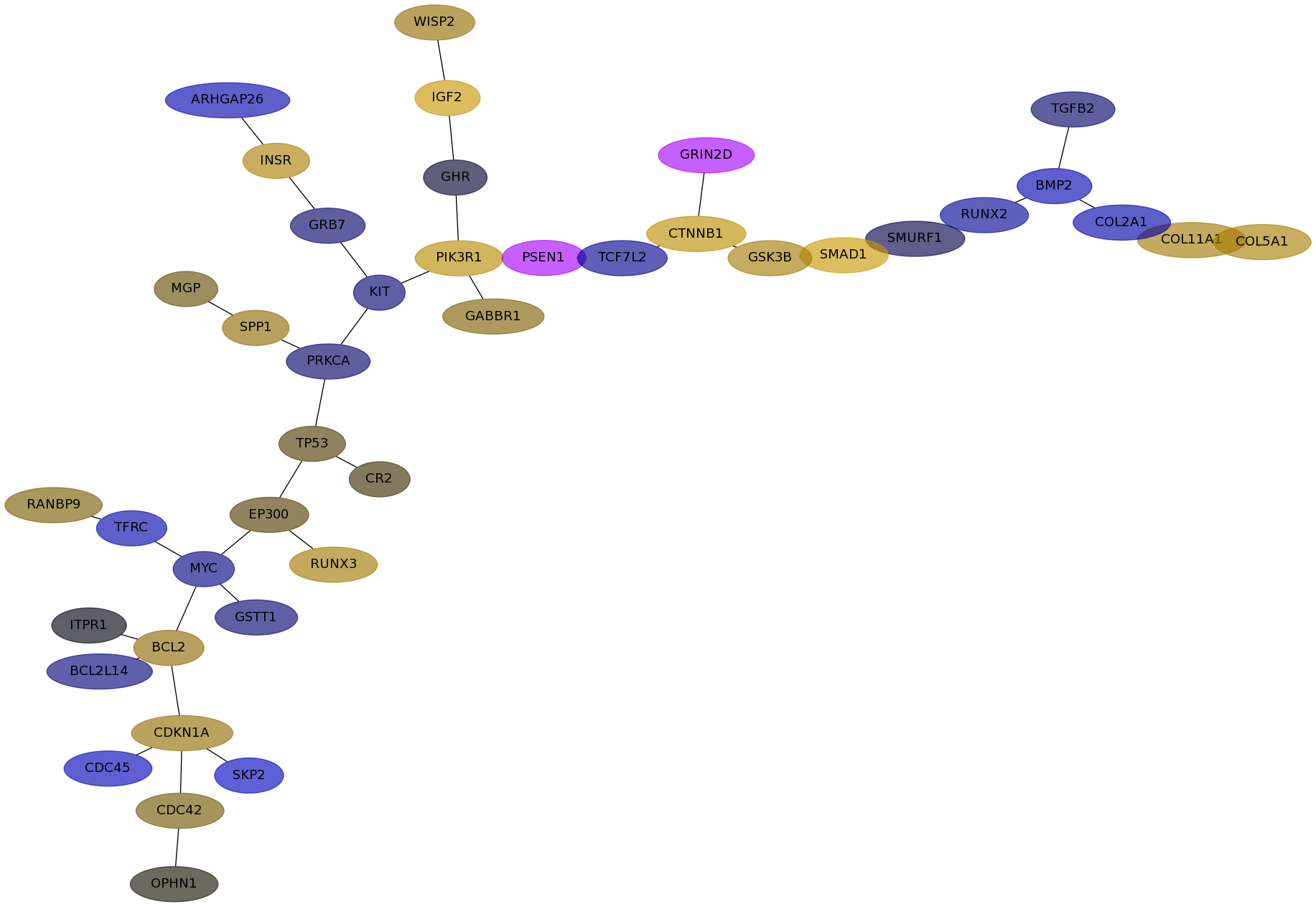
Score for each gene in subnetwork 528 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| ARHGAP26 |   | 5 | 148 | 444 | 440 | -0.145 | -0.023 | 0.171 | -0.222 | -0.348 |
|---|
| RUNX2 |   | 2 | 391 | 364 | 375 | -0.087 | 0.184 | -0.104 | -0.127 | undef |
|---|
| MGP |   | 1 | 652 | 847 | 865 | 0.026 | -0.088 | 0.007 | 0.150 | -0.030 |
|---|
| GABBR1 |   | 2 | 391 | 847 | 841 | 0.049 | -0.107 | 0.164 | 0.081 | 0.131 |
|---|
| OPHN1 |   | 5 | 148 | 339 | 337 | 0.003 | 0.062 | 0.278 | 0.194 | -0.182 |
|---|
| TP53 |   | 28 | 15 | 1 | 9 | 0.015 | -0.167 | 0.148 | -0.027 | 0.147 |
|---|
| GSTT1 |   | 6 | 120 | 364 | 355 | -0.034 | 0.148 | 0.107 | -0.190 | -0.064 |
|---|
| COL11A1 |   | 1 | 652 | 847 | 865 | 0.111 | 0.191 | 0.149 | -0.023 | 0.309 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| TGFB2 |   | 7 | 100 | 57 | 60 | -0.028 | -0.063 | 0.244 | 0.226 | 0.015 |
|---|
| PRKCA |   | 5 | 148 | 339 | 337 | -0.029 | -0.016 | -0.088 | 0.174 | 0.005 |
|---|
| PSEN1 |   | 5 | 148 | 847 | 821 | undef | -0.041 | 0.110 | undef | undef |
|---|
| TFRC |   | 1 | 652 | 847 | 865 | -0.140 | 0.038 | -0.066 | 0.076 | -0.014 |
|---|
| WISP2 |   | 1 | 652 | 847 | 865 | 0.075 | 0.232 | -0.010 | 0.088 | 0.204 |
|---|
| BMP2 |   | 7 | 100 | 33 | 37 | -0.155 | -0.011 | 0.097 | 0.148 | -0.069 |
|---|
| BCL2L14 |   | 1 | 652 | 847 | 865 | -0.044 | -0.113 | -0.224 | 0.118 | -0.238 |
|---|
| SPP1 |   | 4 | 198 | 339 | 341 | 0.068 | 0.130 | 0.119 | 0.184 | 0.153 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| MYC |   | 24 | 21 | 148 | 137 | -0.056 | -0.101 | -0.169 | 0.188 | -0.134 |
|---|
| GHR |   | 14 | 42 | 148 | 146 | -0.006 | -0.112 | 0.174 | 0.090 | 0.201 |
|---|
| COL5A1 |   | 1 | 652 | 847 | 865 | 0.125 | 0.016 | 0.226 | 0.006 | 0.201 |
|---|
| COL2A1 |   | 1 | 652 | 847 | 865 | -0.127 | -0.077 | -0.000 | 0.174 | 0.173 |
|---|
| BCL2 |   | 11 | 59 | 364 | 350 | 0.067 | -0.077 | -0.035 | 0.128 | 0.043 |
|---|
| KIT |   | 13 | 47 | 339 | 335 | -0.034 | -0.130 | -0.091 | 0.235 | -0.059 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| SKP2 |   | 14 | 42 | 148 | 146 | -0.228 | 0.092 | -0.043 | 0.237 | -0.009 |
|---|
| CR2 |   | 2 | 391 | 847 | 841 | 0.010 | -0.042 | -0.089 | 0.101 | 0.015 |
|---|
| RUNX3 |   | 8 | 84 | 33 | 35 | 0.109 | 0.071 | -0.097 | 0.083 | -0.074 |
|---|
| RANBP9 |   | 1 | 652 | 847 | 865 | 0.045 | 0.028 | 0.186 | 0.063 | 0.116 |
|---|
| TCF7L2 |   | 17 | 30 | 271 | 264 | -0.071 | -0.021 | 0.143 | 0.280 | 0.101 |
|---|
| CDC45 |   | 4 | 198 | 847 | 830 | -0.177 | 0.134 | -0.171 | 0.185 | -0.085 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| GRB7 |   | 13 | 47 | 57 | 51 | -0.028 | 0.021 | 0.115 | -0.072 | 0.129 |
|---|
| ITPR1 |   | 3 | 265 | 182 | 189 | -0.002 | -0.096 | 0.077 | 0.015 | 0.008 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| GRIN2D |   | 18 | 26 | 33 | 32 | undef | 0.058 | 0.155 | 0.000 | undef |
|---|
| SMAD1 |   | 24 | 21 | 33 | 30 | 0.287 | 0.049 | 0.159 | -0.062 | 0.048 |
|---|
| SMURF1 |   | 9 | 77 | 33 | 34 | -0.013 | -0.222 | 0.236 | -0.049 | -0.006 |
|---|
| IGF2 |   | 6 | 120 | 532 | 514 | 0.261 | -0.022 | 0.172 | 0.145 | 0.116 |
|---|
GO Enrichment output for subnetwork 528 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of ossification | GO:0030278 |  | 8.543E-10 | 1.965E-06 |
|---|
| cartilage development | GO:0051216 |  | 1.357E-09 | 1.561E-06 |
|---|
| regulation of bone remodeling | GO:0046850 |  | 2.403E-09 | 1.842E-06 |
|---|
| regulation of tissue remodeling | GO:0034103 |  | 2.752E-09 | 1.582E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.602E-08 | 7.371E-06 |
|---|
| collagen fibril organization | GO:0030199 |  | 1.748E-08 | 6.7E-06 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 2.044E-08 | 6.716E-06 |
|---|
| chondrocyte differentiation | GO:0002062 |  | 9.543E-08 | 2.744E-05 |
|---|
| cell fate commitment | GO:0045165 |  | 1.198E-07 | 3.062E-05 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 1.291E-07 | 2.97E-05 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.867E-07 | 5.995E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.166E-12 | 2.85E-09 |
|---|
| regulation of ossification | GO:0030278 |  | 8.063E-10 | 9.849E-07 |
|---|
| cartilage development | GO:0051216 |  | 1.233E-09 | 1.004E-06 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 2.058E-09 | 1.257E-06 |
|---|
| regulation of bone remodeling | GO:0046850 |  | 2.088E-09 | 1.02E-06 |
|---|
| regulation of tissue remodeling | GO:0034103 |  | 2.367E-09 | 9.638E-07 |
|---|
| mesenchymal cell development | GO:0014031 |  | 1.035E-08 | 3.611E-06 |
|---|
| collagen fibril organization | GO:0030199 |  | 1.369E-08 | 4.182E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.851E-08 | 5.025E-06 |
|---|
| osteoblast differentiation | GO:0001649 |  | 4.36E-08 | 1.065E-05 |
|---|
| ossification | GO:0001503 |  | 5.747E-08 | 1.276E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 7.69E-13 | 1.85E-09 |
|---|
| regulation of ossification | GO:0030278 |  | 1.227E-09 | 1.476E-06 |
|---|
| cartilage development | GO:0051216 |  | 1.633E-09 | 1.31E-06 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 2.801E-09 | 1.685E-06 |
|---|
| regulation of bone remodeling | GO:0046850 |  | 3.174E-09 | 1.527E-06 |
|---|
| regulation of tissue remodeling | GO:0034103 |  | 3.597E-09 | 1.443E-06 |
|---|
| mesenchymal cell development | GO:0014031 |  | 1.274E-08 | 4.379E-06 |
|---|
| collagen fibril organization | GO:0030199 |  | 1.862E-08 | 5.599E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.659E-08 | 7.109E-06 |
|---|
| osteoblast differentiation | GO:0001649 |  | 4.798E-08 | 1.154E-05 |
|---|
| chondrocyte differentiation | GO:0002062 |  | 8.004E-08 | 1.751E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of ossification | GO:0030278 |  | 8.543E-10 | 1.965E-06 |
|---|
| cartilage development | GO:0051216 |  | 1.357E-09 | 1.561E-06 |
|---|
| regulation of bone remodeling | GO:0046850 |  | 2.403E-09 | 1.842E-06 |
|---|
| regulation of tissue remodeling | GO:0034103 |  | 2.752E-09 | 1.582E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.602E-08 | 7.371E-06 |
|---|
| collagen fibril organization | GO:0030199 |  | 1.748E-08 | 6.7E-06 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 2.044E-08 | 6.716E-06 |
|---|
| chondrocyte differentiation | GO:0002062 |  | 9.543E-08 | 2.744E-05 |
|---|
| cell fate commitment | GO:0045165 |  | 1.198E-07 | 3.062E-05 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 1.291E-07 | 2.97E-05 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.867E-07 | 5.995E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of ossification | GO:0030278 |  | 8.543E-10 | 1.965E-06 |
|---|
| cartilage development | GO:0051216 |  | 1.357E-09 | 1.561E-06 |
|---|
| regulation of bone remodeling | GO:0046850 |  | 2.403E-09 | 1.842E-06 |
|---|
| regulation of tissue remodeling | GO:0034103 |  | 2.752E-09 | 1.582E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.602E-08 | 7.371E-06 |
|---|
| collagen fibril organization | GO:0030199 |  | 1.748E-08 | 6.7E-06 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 2.044E-08 | 6.716E-06 |
|---|
| chondrocyte differentiation | GO:0002062 |  | 9.543E-08 | 2.744E-05 |
|---|
| cell fate commitment | GO:0045165 |  | 1.198E-07 | 3.062E-05 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 1.291E-07 | 2.97E-05 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.867E-07 | 5.995E-05 |
|---|


