Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1245
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3342 | 1.448e-03 | 2.500e-05 | 1.596e-01 | 5.776e-09 |
|---|
| IPC | 0.3301 | 3.664e-01 | 1.941e-01 | 9.393e-01 | 6.679e-02 |
|---|
| Loi | 0.4862 | 3.500e-05 | 0.000e+00 | 4.864e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5996 | 1.000e-06 | 0.000e+00 | 1.136e-01 | 0.000e+00 |
|---|
| Wang | 0.2554 | 8.261e-03 | 6.961e-02 | 2.383e-01 | 1.370e-04 |
|---|
Expression data for subnetwork 1245 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
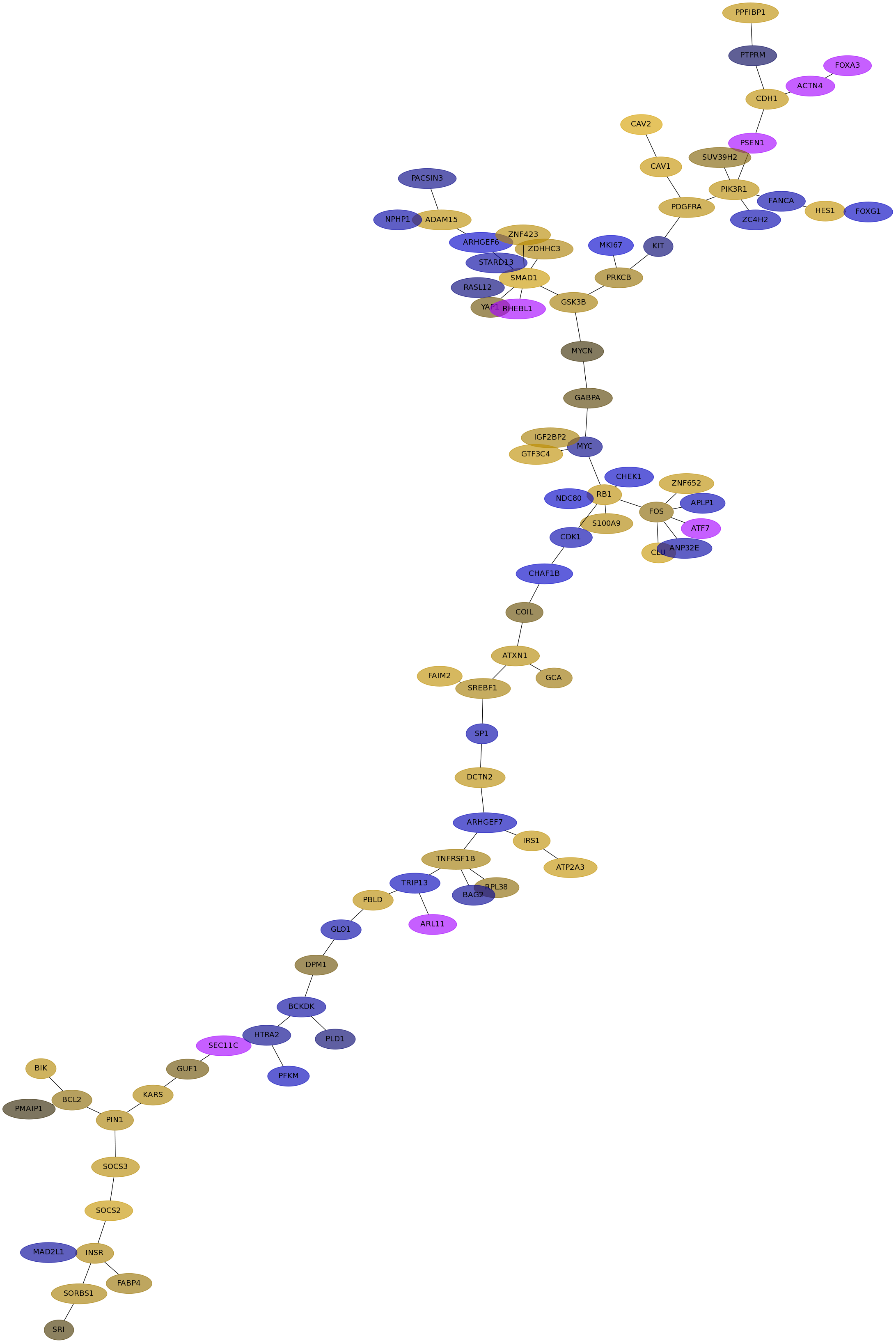
Score for each gene in subnetwork 1245 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| KARS |   | 1 | 652 | 859 | 875 | 0.146 | 0.015 | 0.145 | 0.041 | 0.083 |
|---|
| SORBS1 |   | 17 | 30 | 182 | 178 | 0.124 | -0.043 | 0.015 | 0.314 | 0.200 |
|---|
| SOCS2 |   | 4 | 198 | 859 | 837 | 0.262 | -0.130 | -0.028 | 0.051 | 0.027 |
|---|
| NDC80 |   | 2 | 391 | 57 | 80 | -0.266 | 0.246 | -0.229 | 0.060 | 0.044 |
|---|
| PDGFRA |   | 1 | 652 | 859 | 875 | 0.179 | -0.035 | 0.008 | 0.118 | 0.119 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| PMAIP1 |   | 1 | 652 | 859 | 875 | 0.007 | 0.118 | -0.105 | 0.057 | -0.047 |
|---|
| PPFIBP1 |   | 2 | 391 | 859 | 860 | 0.212 | -0.073 | 0.207 | -0.072 | -0.016 |
|---|
| PIN1 |   | 17 | 30 | 1 | 13 | 0.135 | 0.194 | 0.040 | -0.040 | -0.048 |
|---|
| ARL11 |   | 1 | 652 | 859 | 875 | undef | 0.150 | undef | undef | undef |
|---|
| SRI |   | 5 | 148 | 563 | 553 | 0.013 | 0.270 | -0.040 | 0.181 | 0.057 |
|---|
| KIT |   | 13 | 47 | 339 | 335 | -0.034 | -0.130 | -0.091 | 0.235 | -0.059 |
|---|
| TRIP13 |   | 1 | 652 | 859 | 875 | -0.185 | 0.043 | -0.199 | -0.017 | -0.146 |
|---|
| HTRA2 |   | 2 | 391 | 859 | 860 | -0.063 | -0.123 | -0.118 | 0.234 | 0.011 |
|---|
| GCA |   | 2 | 391 | 419 | 433 | 0.092 | 0.022 | -0.143 | -0.202 | 0.180 |
|---|
| GABPA |   | 6 | 120 | 206 | 200 | 0.019 | 0.033 | -0.183 | 0.066 | -0.161 |
|---|
| DPM1 |   | 1 | 652 | 859 | 875 | 0.029 | 0.177 | -0.072 | 0.133 | 0.009 |
|---|
| PTPRM |   | 1 | 652 | 859 | 875 | -0.019 | -0.132 | 0.049 | 0.101 | 0.066 |
|---|
| RPL38 |   | 3 | 265 | 859 | 843 | 0.065 | 0.222 | 0.151 | 0.241 | -0.171 |
|---|
| MKI67 |   | 3 | 265 | 778 | 770 | -0.295 | 0.205 | -0.123 | 0.176 | 0.018 |
|---|
| YAP1 |   | 2 | 391 | 665 | 660 | 0.030 | -0.199 | 0.132 | 0.262 | 0.075 |
|---|
| ATXN1 |   | 6 | 120 | 364 | 355 | 0.171 | 0.026 | 0.192 | 0.175 | 0.163 |
|---|
| GLO1 |   | 3 | 265 | 859 | 843 | -0.123 | 0.266 | -0.089 | 0.135 | 0.076 |
|---|
| CAV1 |   | 10 | 67 | 439 | 428 | 0.251 | -0.092 | 0.124 | 0.079 | 0.119 |
|---|
| CDK1 |   | 16 | 34 | 206 | 194 | -0.138 | 0.265 | -0.195 | 0.106 | 0.031 |
|---|
| SOCS3 |   | 3 | 265 | 492 | 503 | 0.180 | 0.201 | -0.110 | 0.064 | -0.138 |
|---|
| ARHGEF6 |   | 3 | 265 | 859 | 843 | -0.274 | -0.068 | -0.120 | 0.166 | -0.115 |
|---|
| PRKCB |   | 8 | 84 | 33 | 35 | 0.081 | -0.065 | 0.001 | -0.148 | -0.035 |
|---|
| MAD2L1 |   | 2 | 391 | 57 | 80 | -0.089 | 0.208 | -0.130 | 0.116 | 0.063 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| ACTN4 |   | 6 | 120 | 859 | 834 | undef | 0.035 | 0.180 | undef | undef |
|---|
| ANP32E |   | 6 | 120 | 148 | 156 | -0.114 | 0.166 | -0.139 | 0.191 | 0.118 |
|---|
| CLU |   | 10 | 67 | 117 | 121 | 0.280 | -0.208 | 0.119 | -0.153 | -0.234 |
|---|
| SMAD1 |   | 24 | 21 | 33 | 30 | 0.287 | 0.049 | 0.159 | -0.062 | 0.048 |
|---|
| STARD13 |   | 2 | 391 | 859 | 860 | -0.108 | -0.083 | 0.006 | 0.126 | 0.028 |
|---|
| COIL |   | 3 | 265 | 584 | 573 | 0.027 | 0.151 | 0.144 | 0.071 | 0.044 |
|---|
| RHEBL1 |   | 2 | 391 | 492 | 506 | undef | 0.154 | undef | undef | undef |
|---|
| FABP4 |   | 3 | 265 | 444 | 447 | 0.091 | -0.036 | -0.028 | 0.227 | 0.107 |
|---|
| BCKDK |   | 2 | 391 | 492 | 506 | -0.100 | 0.166 | -0.004 | -0.012 | -0.001 |
|---|
| FAIM2 |   | 2 | 391 | 859 | 860 | 0.220 | 0.056 | 0.001 | -0.159 | -0.203 |
|---|
| ATP2A3 |   | 9 | 77 | 1 | 18 | 0.240 | -0.040 | 0.082 | -0.229 | -0.025 |
|---|
| ARHGEF7 |   | 12 | 55 | 492 | 475 | -0.180 | -0.074 | -0.061 | 0.208 | 0.015 |
|---|
| ATF7 |   | 1 | 652 | 859 | 875 | undef | 0.085 | 0.115 | -0.088 | -0.092 |
|---|
| BIK |   | 2 | 391 | 665 | 660 | 0.172 | -0.097 | 0.133 | -0.179 | -0.071 |
|---|
| DCTN2 |   | 3 | 265 | 859 | 843 | 0.192 | 0.014 | 0.150 | 0.061 | 0.303 |
|---|
| FOXA3 |   | 6 | 120 | 657 | 631 | undef | 0.192 | 0.289 | undef | undef |
|---|
| IGF2BP2 |   | 3 | 265 | 148 | 166 | 0.124 | 0.210 | -0.002 | 0.098 | -0.122 |
|---|
| CHEK1 |   | 1 | 652 | 859 | 875 | -0.240 | 0.189 | -0.134 | 0.119 | -0.054 |
|---|
| BAG2 |   | 2 | 391 | 364 | 375 | -0.081 | 0.016 | 0.178 | 0.152 | 0.083 |
|---|
| FOXG1 |   | 1 | 652 | 859 | 875 | -0.227 | 0.136 | -0.156 | -0.126 | -0.068 |
|---|
| PSEN1 |   | 5 | 148 | 847 | 821 | undef | -0.041 | 0.110 | undef | undef |
|---|
| APLP1 |   | 3 | 265 | 859 | 843 | -0.170 | 0.284 | 0.043 | -0.105 | -0.190 |
|---|
| RB1 |   | 26 | 18 | 271 | 263 | 0.200 | 0.013 | -0.051 | 0.057 | -0.082 |
|---|
| SUV39H2 |   | 3 | 265 | 859 | 843 | 0.049 | 0.150 | -0.097 | -0.057 | 0.028 |
|---|
| MYC |   | 24 | 21 | 148 | 137 | -0.056 | -0.101 | -0.169 | 0.188 | -0.134 |
|---|
| SREBF1 |   | 2 | 391 | 859 | 860 | 0.119 | -0.042 | 0.232 | -0.195 | -0.074 |
|---|
| ADAM15 |   | 13 | 47 | 492 | 474 | 0.205 | -0.048 | -0.078 | -0.208 | -0.048 |
|---|
| BCL2 |   | 11 | 59 | 364 | 350 | 0.067 | -0.077 | -0.035 | 0.128 | 0.043 |
|---|
| MYCN |   | 3 | 265 | 859 | 843 | 0.009 | 0.125 | 0.057 | -0.003 | -0.084 |
|---|
| ZNF423 |   | 1 | 652 | 859 | 875 | 0.192 | -0.061 | 0.100 | 0.017 | 0.205 |
|---|
| S100A9 |   | 18 | 26 | 398 | 382 | 0.154 | 0.117 | 0.043 | -0.110 | -0.045 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| SP1 |   | 16 | 34 | 283 | 272 | -0.129 | 0.222 | -0.254 | 0.239 | 0.082 |
|---|
| RASL12 |   | 3 | 265 | 665 | 651 | -0.041 | -0.154 | 0.210 | 0.152 | 0.004 |
|---|
| HES1 |   | 4 | 198 | 746 | 725 | 0.247 | 0.115 | 0.157 | 0.010 | -0.106 |
|---|
| CHAF1B |   | 2 | 391 | 584 | 578 | -0.264 | 0.015 | -0.193 | 0.171 | -0.041 |
|---|
| ZC4H2 |   | 16 | 34 | 339 | 332 | -0.145 | 0.065 | 0.061 | 0.304 | 0.059 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| PACSIN3 |   | 3 | 265 | 778 | 770 | -0.055 | 0.270 | 0.129 | -0.045 | 0.066 |
|---|
| SEC11C |   | 1 | 652 | 859 | 875 | undef | 0.141 | -0.215 | undef | undef |
|---|
| GUF1 |   | 1 | 652 | 859 | 875 | 0.030 | 0.099 | -0.059 | 0.058 | -0.111 |
|---|
| PBLD |   | 1 | 652 | 859 | 875 | 0.195 | -0.082 | 0.095 | 0.030 | 0.086 |
|---|
| FOS |   | 20 | 24 | 148 | 138 | 0.060 | -0.137 | 0.129 | 0.185 | 0.234 |
|---|
| FANCA |   | 3 | 265 | 57 | 71 | -0.131 | 0.120 | -0.188 | 0.106 | -0.053 |
|---|
| PFKM |   | 4 | 198 | 492 | 499 | -0.193 | -0.030 | -0.079 | 0.293 | 0.118 |
|---|
| ZDHHC3 |   | 1 | 652 | 859 | 875 | 0.138 | -0.193 | 0.075 | -0.014 | -0.023 |
|---|
| TNFRSF1B |   | 12 | 55 | 563 | 544 | 0.109 | 0.024 | -0.187 | -0.096 | -0.106 |
|---|
| IRS1 |   | 14 | 42 | 1 | 14 | 0.217 | -0.219 | 0.065 | 0.165 | 0.064 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| GTF3C4 |   | 1 | 652 | 859 | 875 | 0.221 | 0.024 | 0.074 | 0.235 | -0.147 |
|---|
| ZNF652 |   | 3 | 265 | 536 | 537 | 0.208 | -0.215 | -0.045 | -0.150 | -0.076 |
|---|
| PLD1 |   | 2 | 391 | 492 | 506 | -0.031 | 0.132 | 0.033 | -0.158 | 0.123 |
|---|
| NPHP1 |   | 2 | 391 | 206 | 220 | -0.113 | 0.139 | 0.095 | 0.026 | -0.110 |
|---|
GO Enrichment output for subnetwork 1245 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 2.77E-07 | 6.371E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 5.826E-07 | 6.7E-04 |
|---|
| gland morphogenesis | GO:0022612 |  | 1.07E-06 | 8.205E-04 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.141E-06 | 1.231E-03 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 2.769E-06 | 1.274E-03 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 3.282E-06 | 1.258E-03 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 3.282E-06 | 1.078E-03 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 3.607E-06 | 1.037E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 6.024E-06 | 1.539E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 6.405E-06 | 1.473E-03 |
|---|
| gas homeostasis | GO:0033483 |  | 6.531E-06 | 1.366E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 4.319E-07 | 1.055E-03 |
|---|
| gland morphogenesis | GO:0022612 |  | 7.234E-07 | 8.836E-04 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.008E-06 | 8.21E-04 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 1.847E-06 | 1.128E-03 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 1.887E-06 | 9.219E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 2.069E-06 | 8.424E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 2.551E-06 | 8.904E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 4.75E-06 | 1.45E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 5.841E-06 | 1.585E-03 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 6.411E-06 | 1.566E-03 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 6.411E-06 | 1.424E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 4.169E-07 | 1.003E-03 |
|---|
| gland morphogenesis | GO:0022612 |  | 5.755E-07 | 6.923E-04 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.153E-06 | 9.247E-04 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 1.716E-06 | 1.032E-03 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 2.047E-06 | 9.848E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 2.283E-06 | 9.155E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 2.62E-06 | 9.005E-04 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 4.075E-06 | 1.226E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 5.427E-06 | 1.451E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 5.427E-06 | 1.306E-03 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 7.101E-06 | 1.553E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 2.77E-07 | 6.371E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 5.826E-07 | 6.7E-04 |
|---|
| gland morphogenesis | GO:0022612 |  | 1.07E-06 | 8.205E-04 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.141E-06 | 1.231E-03 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 2.769E-06 | 1.274E-03 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 3.282E-06 | 1.258E-03 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 3.282E-06 | 1.078E-03 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 3.607E-06 | 1.037E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 6.024E-06 | 1.539E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 6.405E-06 | 1.473E-03 |
|---|
| gas homeostasis | GO:0033483 |  | 6.531E-06 | 1.366E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 2.77E-07 | 6.371E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 5.826E-07 | 6.7E-04 |
|---|
| gland morphogenesis | GO:0022612 |  | 1.07E-06 | 8.205E-04 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.141E-06 | 1.231E-03 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 2.769E-06 | 1.274E-03 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 3.282E-06 | 1.258E-03 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 3.282E-06 | 1.078E-03 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 3.607E-06 | 1.037E-03 |
|---|
| protein oligomerization | GO:0051259 |  | 6.024E-06 | 1.539E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 6.405E-06 | 1.473E-03 |
|---|
| gas homeostasis | GO:0033483 |  | 6.531E-06 | 1.366E-03 |
|---|


