Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1218
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3324 | 1.574e-03 | 2.800e-05 | 1.654e-01 | 7.289e-09 |
|---|
| IPC | 0.3393 | 3.514e-01 | 1.807e-01 | 9.362e-01 | 5.943e-02 |
|---|
| Loi | 0.4837 | 3.900e-05 | 0.000e+00 | 4.983e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6019 | 1.000e-06 | 0.000e+00 | 1.090e-01 | 0.000e+00 |
|---|
| Wang | 0.2570 | 7.733e-03 | 6.680e-02 | 2.311e-01 | 1.194e-04 |
|---|
Expression data for subnetwork 1218 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
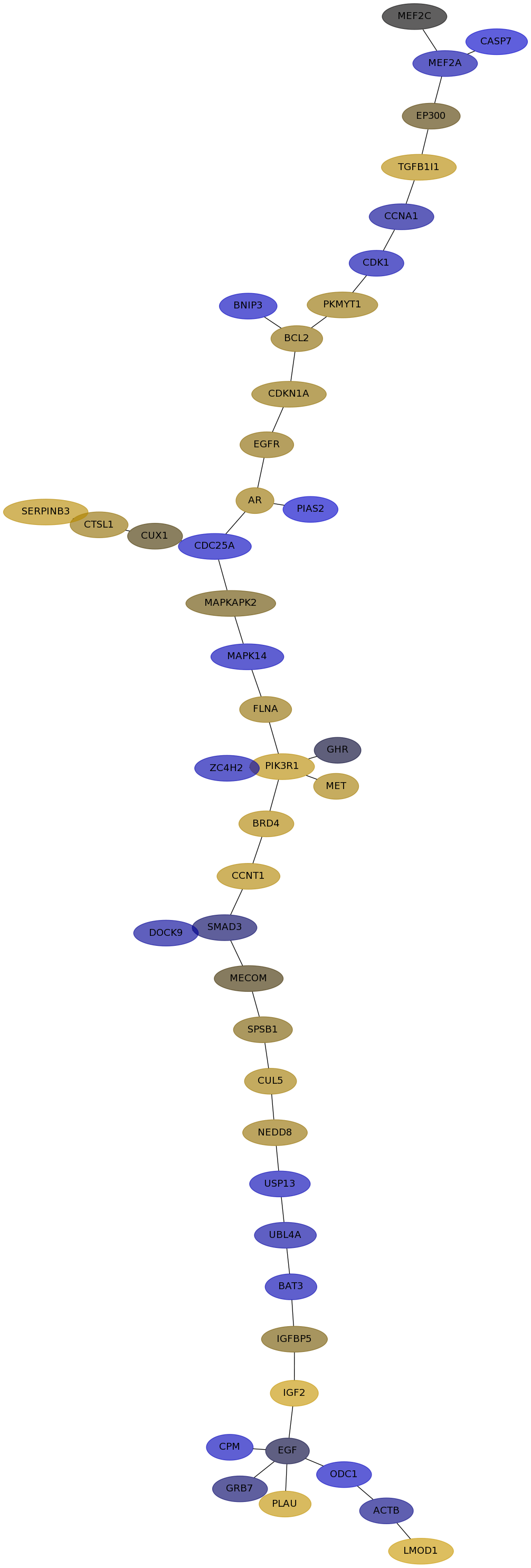
Score for each gene in subnetwork 1218 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| UBL4A |   | 1 | 652 | 620 | 636 | -0.105 | 0.196 | -0.040 | 0.219 | 0.062 |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| EGF |   | 11 | 59 | 398 | 383 | -0.009 | -0.212 | 0.182 | -0.276 | 0.005 |
|---|
| LMOD1 |   | 1 | 652 | 620 | 636 | 0.287 | -0.086 | 0.183 | 0.175 | 0.118 |
|---|
| MEF2A |   | 5 | 148 | 283 | 287 | -0.123 | 0.022 | 0.036 | 0.265 | 0.124 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| SPSB1 |   | 7 | 100 | 492 | 495 | 0.044 | -0.124 | 0.088 | -0.152 | 0.169 |
|---|
| MAPK14 |   | 2 | 391 | 620 | 618 | -0.183 | 0.033 | -0.065 | 0.299 | 0.119 |
|---|
| MECOM |   | 1 | 652 | 620 | 636 | 0.010 | -0.129 | 0.127 | 0.138 | 0.055 |
|---|
| CUL5 |   | 4 | 198 | 492 | 499 | 0.111 | 0.051 | 0.187 | 0.221 | 0.216 |
|---|
| PLAU |   | 6 | 120 | 444 | 438 | 0.238 | 0.187 | 0.153 | 0.141 | 0.192 |
|---|
| GHR |   | 14 | 42 | 148 | 146 | -0.006 | -0.112 | 0.174 | 0.090 | 0.201 |
|---|
| BCL2 |   | 11 | 59 | 364 | 350 | 0.067 | -0.077 | -0.035 | 0.128 | 0.043 |
|---|
| BNIP3 |   | 1 | 652 | 620 | 636 | -0.207 | 0.193 | 0.071 | 0.102 | 0.285 |
|---|
| MET |   | 5 | 148 | 536 | 536 | 0.119 | -0.219 | 0.086 | 0.183 | 0.170 |
|---|
| BAT3 |   | 2 | 391 | 444 | 458 | -0.153 | -0.078 | 0.072 | -0.057 | 0.069 |
|---|
| CDC25A |   | 5 | 148 | 444 | 440 | -0.220 | 0.113 | -0.170 | 0.151 | -0.003 |
|---|
| SERPINB3 |   | 1 | 652 | 620 | 636 | 0.190 | -0.113 | 0.084 | 0.221 | 0.006 |
|---|
| BRD4 |   | 18 | 26 | 1 | 12 | 0.156 | -0.119 | 0.095 | -0.075 | -0.044 |
|---|
| ZC4H2 |   | 16 | 34 | 339 | 332 | -0.145 | 0.065 | 0.061 | 0.304 | 0.059 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| CUX1 |   | 8 | 84 | 271 | 266 | 0.012 | -0.042 | 0.256 | -0.121 | 0.040 |
|---|
| PKMYT1 |   | 5 | 148 | 620 | 603 | 0.085 | 0.175 | -0.143 | 0.046 | -0.070 |
|---|
| FLNA |   | 7 | 100 | 206 | 198 | 0.077 | 0.081 | 0.107 | 0.148 | 0.126 |
|---|
| ODC1 |   | 5 | 148 | 398 | 387 | -0.220 | -0.006 | -0.099 | 0.209 | -0.107 |
|---|
| CDK1 |   | 16 | 34 | 206 | 194 | -0.138 | 0.265 | -0.195 | 0.106 | 0.031 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| MAPKAPK2 |   | 1 | 652 | 620 | 636 | 0.028 | 0.149 | 0.107 | 0.141 | 0.009 |
|---|
| CTSL1 |   | 3 | 265 | 283 | 292 | 0.076 | 0.187 | -0.080 | 0.091 | 0.178 |
|---|
| NEDD8 |   | 3 | 265 | 536 | 537 | 0.083 | 0.250 | -0.085 | 0.251 | -0.073 |
|---|
| SMAD3 |   | 1 | 652 | 620 | 636 | -0.024 | -0.176 | 0.063 | 0.239 | -0.012 |
|---|
| CASP7 |   | 8 | 84 | 283 | 278 | -0.270 | 0.216 | 0.095 | 0.255 | 0.035 |
|---|
| CCNT1 |   | 18 | 26 | 33 | 32 | 0.163 | 0.008 | -0.028 | 0.162 | -0.213 |
|---|
| GRB7 |   | 13 | 47 | 57 | 51 | -0.028 | 0.021 | 0.115 | -0.072 | 0.129 |
|---|
| IGFBP5 |   | 3 | 265 | 444 | 447 | 0.038 | -0.076 | 0.110 | 0.074 | 0.181 |
|---|
| CCNA1 |   | 17 | 30 | 326 | 314 | -0.078 | 0.174 | 0.092 | 0.078 | -0.098 |
|---|
| PIAS2 |   | 3 | 265 | 620 | 613 | -0.270 | 0.039 | -0.072 | 0.194 | -0.172 |
|---|
| CPM |   | 2 | 391 | 620 | 618 | -0.196 | 0.166 | -0.062 | -0.079 | 0.280 |
|---|
| AR |   | 9 | 77 | 283 | 277 | 0.089 | -0.163 | 0.081 | -0.035 | -0.030 |
|---|
| ACTB |   | 7 | 100 | 398 | 385 | -0.052 | -0.039 | 0.283 | -0.061 | 0.003 |
|---|
| IGF2 |   | 6 | 120 | 532 | 514 | 0.261 | -0.022 | 0.172 | 0.145 | 0.116 |
|---|
| TGFB1I1 |   | 16 | 34 | 326 | 315 | 0.176 | -0.105 | 0.229 | 0.143 | 0.298 |
|---|
| DOCK9 |   | 1 | 652 | 620 | 636 | -0.086 | -0.021 | 0.128 | -0.023 | -0.016 |
|---|
| MEF2C |   | 1 | 652 | 620 | 636 | 0.000 | -0.062 | -0.097 | 0.112 | 0.132 |
|---|
| USP13 |   | 1 | 652 | 620 | 636 | -0.169 | -0.004 | -0.268 | 0.127 | 0.055 |
|---|
GO Enrichment output for subnetwork 1218 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| neutrophil homeostasis | GO:0001780 |  | 2.545E-11 | 5.854E-08 |
|---|
| pericardium development | GO:0060039 |  | 8.87E-11 | 1.02E-07 |
|---|
| myeloid cell homeostasis | GO:0002262 |  | 2.355E-10 | 1.806E-07 |
|---|
| leukocyte homeostasis | GO:0001776 |  | 1.526E-09 | 8.777E-07 |
|---|
| post-embryonic development | GO:0009791 |  | 4.628E-09 | 2.129E-06 |
|---|
| embryonic hindlimb morphogenesis | GO:0035116 |  | 1.226E-08 | 4.698E-06 |
|---|
| embryonic forelimb morphogenesis | GO:0035115 |  | 1.226E-08 | 4.027E-06 |
|---|
| forelimb morphogenesis | GO:0035136 |  | 2.504E-08 | 7.198E-06 |
|---|
| hindlimb morphogenesis | GO:0035137 |  | 6.193E-08 | 1.583E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.246E-07 | 7.465E-05 |
|---|
| homeostasis of number of cells | GO:0048872 |  | 9.58E-07 | 2.003E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| neutrophil homeostasis | GO:0001780 |  | 6.993E-12 | 1.708E-08 |
|---|
| pericardium development | GO:0060039 |  | 2.439E-11 | 2.98E-08 |
|---|
| myeloid cell homeostasis | GO:0002262 |  | 6.483E-11 | 5.279E-08 |
|---|
| leukocyte homeostasis | GO:0001776 |  | 7.807E-10 | 4.768E-07 |
|---|
| embryonic hindlimb morphogenesis | GO:0035116 |  | 4.923E-09 | 2.405E-06 |
|---|
| post-embryonic development | GO:0009791 |  | 5.749E-09 | 2.341E-06 |
|---|
| embryonic forelimb morphogenesis | GO:0035115 |  | 6.95E-09 | 2.426E-06 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 6.95E-09 | 2.122E-06 |
|---|
| forelimb morphogenesis | GO:0035136 |  | 1.724E-08 | 4.68E-06 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 2.909E-08 | 7.106E-06 |
|---|
| hindlimb morphogenesis | GO:0035137 |  | 2.909E-08 | 6.46E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| neutrophil homeostasis | GO:0001780 |  | 9.309E-12 | 2.24E-08 |
|---|
| pericardium development | GO:0060039 |  | 3.247E-11 | 3.906E-08 |
|---|
| myeloid cell homeostasis | GO:0002262 |  | 8.627E-11 | 6.919E-08 |
|---|
| leukocyte homeostasis | GO:0001776 |  | 8.347E-10 | 5.021E-07 |
|---|
| embryonic hindlimb morphogenesis | GO:0035116 |  | 4.513E-09 | 2.172E-06 |
|---|
| embryonic forelimb morphogenesis | GO:0035115 |  | 6.541E-09 | 2.623E-06 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 6.541E-09 | 2.248E-06 |
|---|
| post-embryonic development | GO:0009791 |  | 6.648E-09 | 1.999E-06 |
|---|
| forelimb morphogenesis | GO:0035136 |  | 1.723E-08 | 4.606E-06 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 2.289E-08 | 5.507E-06 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 2.994E-08 | 6.548E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| neutrophil homeostasis | GO:0001780 |  | 2.545E-11 | 5.854E-08 |
|---|
| pericardium development | GO:0060039 |  | 8.87E-11 | 1.02E-07 |
|---|
| myeloid cell homeostasis | GO:0002262 |  | 2.355E-10 | 1.806E-07 |
|---|
| leukocyte homeostasis | GO:0001776 |  | 1.526E-09 | 8.777E-07 |
|---|
| post-embryonic development | GO:0009791 |  | 4.628E-09 | 2.129E-06 |
|---|
| embryonic hindlimb morphogenesis | GO:0035116 |  | 1.226E-08 | 4.698E-06 |
|---|
| embryonic forelimb morphogenesis | GO:0035115 |  | 1.226E-08 | 4.027E-06 |
|---|
| forelimb morphogenesis | GO:0035136 |  | 2.504E-08 | 7.198E-06 |
|---|
| hindlimb morphogenesis | GO:0035137 |  | 6.193E-08 | 1.583E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.246E-07 | 7.465E-05 |
|---|
| homeostasis of number of cells | GO:0048872 |  | 9.58E-07 | 2.003E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| neutrophil homeostasis | GO:0001780 |  | 2.545E-11 | 5.854E-08 |
|---|
| pericardium development | GO:0060039 |  | 8.87E-11 | 1.02E-07 |
|---|
| myeloid cell homeostasis | GO:0002262 |  | 2.355E-10 | 1.806E-07 |
|---|
| leukocyte homeostasis | GO:0001776 |  | 1.526E-09 | 8.777E-07 |
|---|
| post-embryonic development | GO:0009791 |  | 4.628E-09 | 2.129E-06 |
|---|
| embryonic hindlimb morphogenesis | GO:0035116 |  | 1.226E-08 | 4.698E-06 |
|---|
| embryonic forelimb morphogenesis | GO:0035115 |  | 1.226E-08 | 4.027E-06 |
|---|
| forelimb morphogenesis | GO:0035136 |  | 2.504E-08 | 7.198E-06 |
|---|
| hindlimb morphogenesis | GO:0035137 |  | 6.193E-08 | 1.583E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.246E-07 | 7.465E-05 |
|---|
| homeostasis of number of cells | GO:0048872 |  | 9.58E-07 | 2.003E-04 |
|---|


