Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1180
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3302 | 1.737e-03 | 3.300e-05 | 1.726e-01 | 9.891e-09 |
|---|
| IPC | 0.3481 | 3.369e-01 | 1.681e-01 | 9.332e-01 | 5.285e-02 |
|---|
| Loi | 0.4788 | 4.900e-05 | 0.000e+00 | 5.219e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6052 | 1.000e-06 | 0.000e+00 | 1.024e-01 | 0.000e+00 |
|---|
| Wang | 0.2576 | 7.538e-03 | 6.574e-02 | 2.284e-01 | 1.132e-04 |
|---|
Expression data for subnetwork 1180 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
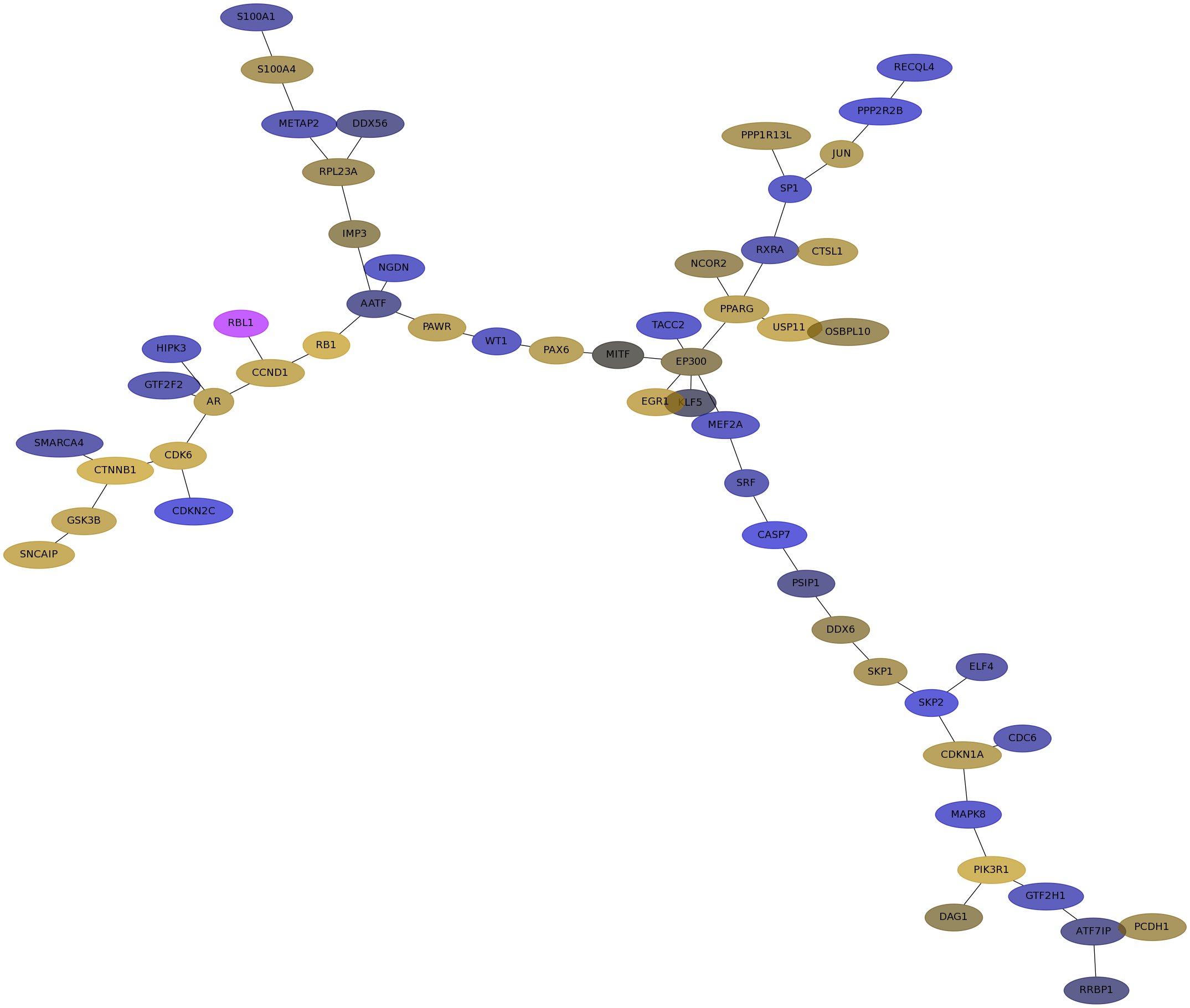
Score for each gene in subnetwork 1180 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| MAPK8 |   | 2 | 391 | 283 | 301 | -0.159 | 0.153 | 0.200 | 0.249 | 0.138 |
|---|
| NCOR2 |   | 11 | 59 | 283 | 274 | 0.024 | -0.096 | 0.211 | -0.163 | 0.114 |
|---|
| PAX6 |   | 11 | 59 | 283 | 274 | 0.081 | 0.259 | -0.003 | 0.112 | 0.168 |
|---|
| EGR1 |   | 11 | 59 | 283 | 274 | 0.118 | -0.184 | 0.259 | 0.130 | 0.122 |
|---|
| RXRA |   | 3 | 265 | 283 | 292 | -0.062 | -0.055 | 0.134 | -0.027 | -0.096 |
|---|
| SMARCA4 |   | 4 | 198 | 283 | 289 | -0.050 | 0.080 | 0.202 | -0.071 | -0.116 |
|---|
| MEF2A |   | 5 | 148 | 283 | 287 | -0.123 | 0.022 | 0.036 | 0.265 | 0.124 |
|---|
| PAWR |   | 1 | 652 | 283 | 318 | 0.088 | 0.173 | 0.224 | 0.034 | 0.137 |
|---|
| ATF7IP |   | 1 | 652 | 283 | 318 | -0.020 | 0.057 | -0.097 | -0.035 | -0.164 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| PSIP1 |   | 1 | 652 | 283 | 318 | -0.019 | 0.081 | -0.276 | 0.220 | 0.126 |
|---|
| RPL23A |   | 10 | 67 | 57 | 52 | 0.031 | 0.208 | -0.007 | 0.024 | 0.055 |
|---|
| JUN |   | 3 | 265 | 283 | 292 | 0.067 | 0.200 | 0.051 | 0.220 | 0.200 |
|---|
| CDKN2C |   | 3 | 265 | 283 | 292 | -0.269 | 0.216 | -0.106 | 0.202 | -0.081 |
|---|
| CCND1 |   | 12 | 55 | 148 | 150 | 0.122 | -0.258 | 0.074 | 0.117 | 0.003 |
|---|
| DDX56 |   | 1 | 652 | 283 | 318 | -0.018 | -0.119 | 0.102 | -0.220 | -0.107 |
|---|
| RB1 |   | 26 | 18 | 271 | 263 | 0.200 | 0.013 | -0.051 | 0.057 | -0.082 |
|---|
| RECQL4 |   | 2 | 391 | 283 | 301 | -0.143 | 0.039 | -0.211 | 0.069 | -0.053 |
|---|
| NGDN |   | 1 | 652 | 283 | 318 | -0.129 | 0.222 | -0.083 | 0.197 | -0.074 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| DDX6 |   | 3 | 265 | 283 | 292 | 0.027 | 0.028 | 0.162 | 0.157 | 0.084 |
|---|
| TACC2 |   | 9 | 77 | 148 | 154 | -0.151 | -0.073 | 0.122 | 0.181 | 0.180 |
|---|
| RRBP1 |   | 1 | 652 | 283 | 318 | -0.014 | -0.094 | 0.216 | -0.200 | 0.137 |
|---|
| MITF |   | 1 | 652 | 283 | 318 | 0.001 | 0.027 | 0.032 | 0.065 | 0.183 |
|---|
| GTF2F2 |   | 3 | 265 | 283 | 292 | -0.060 | -0.059 | -0.085 | 0.119 | -0.054 |
|---|
| WT1 |   | 8 | 84 | 283 | 278 | -0.106 | 0.254 | -0.039 | 0.036 | 0.172 |
|---|
| GTF2H1 |   | 1 | 652 | 283 | 318 | -0.090 | -0.156 | 0.165 | 0.195 | 0.111 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| CDK6 |   | 25 | 19 | 1 | 10 | 0.156 | -0.229 | 0.107 | -0.028 | 0.104 |
|---|
| SP1 |   | 16 | 34 | 283 | 272 | -0.129 | 0.222 | -0.254 | 0.239 | 0.082 |
|---|
| METAP2 |   | 1 | 652 | 283 | 318 | -0.071 | 0.067 | 0.050 | 0.177 | -0.085 |
|---|
| PCDH1 |   | 2 | 391 | 283 | 301 | 0.042 | 0.104 | 0.266 | -0.067 | 0.001 |
|---|
| AATF |   | 1 | 652 | 283 | 318 | -0.020 | 0.056 | -0.163 | 0.053 | -0.036 |
|---|
| PPARG |   | 3 | 265 | 283 | 292 | 0.092 | -0.119 | 0.011 | 0.105 | 0.039 |
|---|
| S100A4 |   | 6 | 120 | 283 | 282 | 0.047 | 0.163 | -0.116 | 0.096 | 0.148 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| SKP2 |   | 14 | 42 | 148 | 146 | -0.228 | 0.092 | -0.043 | 0.237 | -0.009 |
|---|
| CDC6 |   | 4 | 198 | 283 | 289 | -0.059 | 0.198 | -0.053 | 0.038 | 0.095 |
|---|
| OSBPL10 |   | 2 | 391 | 283 | 301 | 0.028 | -0.116 | 0.239 | 0.026 | 0.035 |
|---|
| IMP3 |   | 2 | 391 | 283 | 301 | 0.021 | 0.042 | -0.132 | 0.113 | -0.196 |
|---|
| ELF4 |   | 2 | 391 | 283 | 301 | -0.044 | 0.189 | 0.042 | 0.006 | 0.111 |
|---|
| DAG1 |   | 2 | 391 | 283 | 301 | 0.021 | -0.197 | 0.258 | 0.049 | 0.039 |
|---|
| CTSL1 |   | 3 | 265 | 283 | 292 | 0.076 | 0.187 | -0.080 | 0.091 | 0.178 |
|---|
| CASP7 |   | 8 | 84 | 283 | 278 | -0.270 | 0.216 | 0.095 | 0.255 | 0.035 |
|---|
| RBL1 |   | 1 | 652 | 283 | 318 | undef | 0.078 | undef | undef | undef |
|---|
| S100A1 |   | 3 | 265 | 283 | 292 | -0.045 | 0.244 | 0.044 | 0.013 | 0.063 |
|---|
| AR |   | 9 | 77 | 283 | 277 | 0.089 | -0.163 | 0.081 | -0.035 | -0.030 |
|---|
| SKP1 |   | 1 | 652 | 283 | 318 | 0.048 | 0.143 | 0.037 | 0.055 | -0.110 |
|---|
| SRF |   | 3 | 265 | 283 | 292 | -0.061 | -0.017 | -0.023 | 0.309 | 0.108 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| KLF5 |   | 25 | 19 | 57 | 50 | -0.005 | 0.077 | 0.117 | -0.007 | -0.006 |
|---|
| PPP1R13L |   | 2 | 391 | 283 | 301 | 0.049 | 0.032 | 0.351 | -0.104 | 0.173 |
|---|
| USP11 |   | 5 | 148 | 283 | 287 | 0.134 | -0.144 | 0.106 | 0.121 | -0.044 |
|---|
| SNCAIP |   | 4 | 198 | 283 | 289 | 0.127 | -0.091 | 0.013 | 0.092 | 0.059 |
|---|
| HIPK3 |   | 2 | 391 | 283 | 301 | -0.103 | 0.069 | 0.005 | -0.027 | 0.175 |
|---|
GO Enrichment output for subnetwork 1180 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.928E-08 | 6.735E-05 |
|---|
| interphase | GO:0051325 |  | 3.515E-08 | 4.042E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.456E-07 | 1.117E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.148E-07 | 2.385E-04 |
|---|
| G1 phase | GO:0051318 |  | 1.671E-06 | 7.688E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 7.888E-06 | 3.024E-03 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 9.664E-06 | 3.175E-03 |
|---|
| regulation of transcription initiation from RNA polymerase II promoter | GO:0060260 |  | 9.664E-06 | 2.778E-03 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.117E-05 | 2.855E-03 |
|---|
| detection of bacterium | GO:0016045 |  | 2.054E-05 | 4.724E-03 |
|---|
| N-terminal protein amino acid modification | GO:0031365 |  | 4.835E-05 | 0.01010945 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 9.516E-09 | 2.325E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.163E-08 | 5.085E-05 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 9.826E-08 | 8.002E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.552E-07 | 9.48E-05 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 4.011E-07 | 1.96E-04 |
|---|
| G1 phase | GO:0051318 |  | 2.101E-06 | 8.553E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 3.032E-06 | 1.058E-03 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 4.062E-06 | 1.24E-03 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 4.237E-06 | 1.15E-03 |
|---|
| regulation of transcription initiation from RNA polymerase II promoter | GO:0060260 |  | 6.074E-06 | 1.484E-03 |
|---|
| intracellular receptor-mediated signaling pathway | GO:0030522 |  | 1.302E-05 | 2.892E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.158E-08 | 2.786E-05 |
|---|
| interphase | GO:0051325 |  | 1.483E-08 | 1.783E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.864E-08 | 4.703E-05 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 7.94E-08 | 4.776E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.182E-07 | 1.05E-04 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 5.072E-07 | 2.034E-04 |
|---|
| G1 phase | GO:0051318 |  | 1.42E-06 | 4.882E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 3.827E-06 | 1.151E-03 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 5.347E-06 | 1.429E-03 |
|---|
| regulation of transcription initiation from RNA polymerase II promoter | GO:0060260 |  | 7.254E-06 | 1.745E-03 |
|---|
| intracellular receptor-mediated signaling pathway | GO:0030522 |  | 1.396E-05 | 3.053E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.928E-08 | 6.735E-05 |
|---|
| interphase | GO:0051325 |  | 3.515E-08 | 4.042E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.456E-07 | 1.117E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.148E-07 | 2.385E-04 |
|---|
| G1 phase | GO:0051318 |  | 1.671E-06 | 7.688E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 7.888E-06 | 3.024E-03 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 9.664E-06 | 3.175E-03 |
|---|
| regulation of transcription initiation from RNA polymerase II promoter | GO:0060260 |  | 9.664E-06 | 2.778E-03 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.117E-05 | 2.855E-03 |
|---|
| detection of bacterium | GO:0016045 |  | 2.054E-05 | 4.724E-03 |
|---|
| N-terminal protein amino acid modification | GO:0031365 |  | 4.835E-05 | 0.01010945 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.928E-08 | 6.735E-05 |
|---|
| interphase | GO:0051325 |  | 3.515E-08 | 4.042E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.456E-07 | 1.117E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.148E-07 | 2.385E-04 |
|---|
| G1 phase | GO:0051318 |  | 1.671E-06 | 7.688E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 7.888E-06 | 3.024E-03 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 9.664E-06 | 3.175E-03 |
|---|
| regulation of transcription initiation from RNA polymerase II promoter | GO:0060260 |  | 9.664E-06 | 2.778E-03 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.117E-05 | 2.855E-03 |
|---|
| detection of bacterium | GO:0016045 |  | 2.054E-05 | 4.724E-03 |
|---|
| N-terminal protein amino acid modification | GO:0031365 |  | 4.835E-05 | 0.01010945 |
|---|


