Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1169
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3312 | 1.659e-03 | 3.100e-05 | 1.692e-01 | 8.699e-09 |
|---|
| IPC | 0.3453 | 3.415e-01 | 1.721e-01 | 9.341e-01 | 5.488e-02 |
|---|
| Loi | 0.4780 | 5.100e-05 | 0.000e+00 | 5.259e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6032 | 1.000e-06 | 0.000e+00 | 1.063e-01 | 0.000e+00 |
|---|
| Wang | 0.2568 | 7.805e-03 | 6.719e-02 | 2.321e-01 | 1.217e-04 |
|---|
Expression data for subnetwork 1169 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
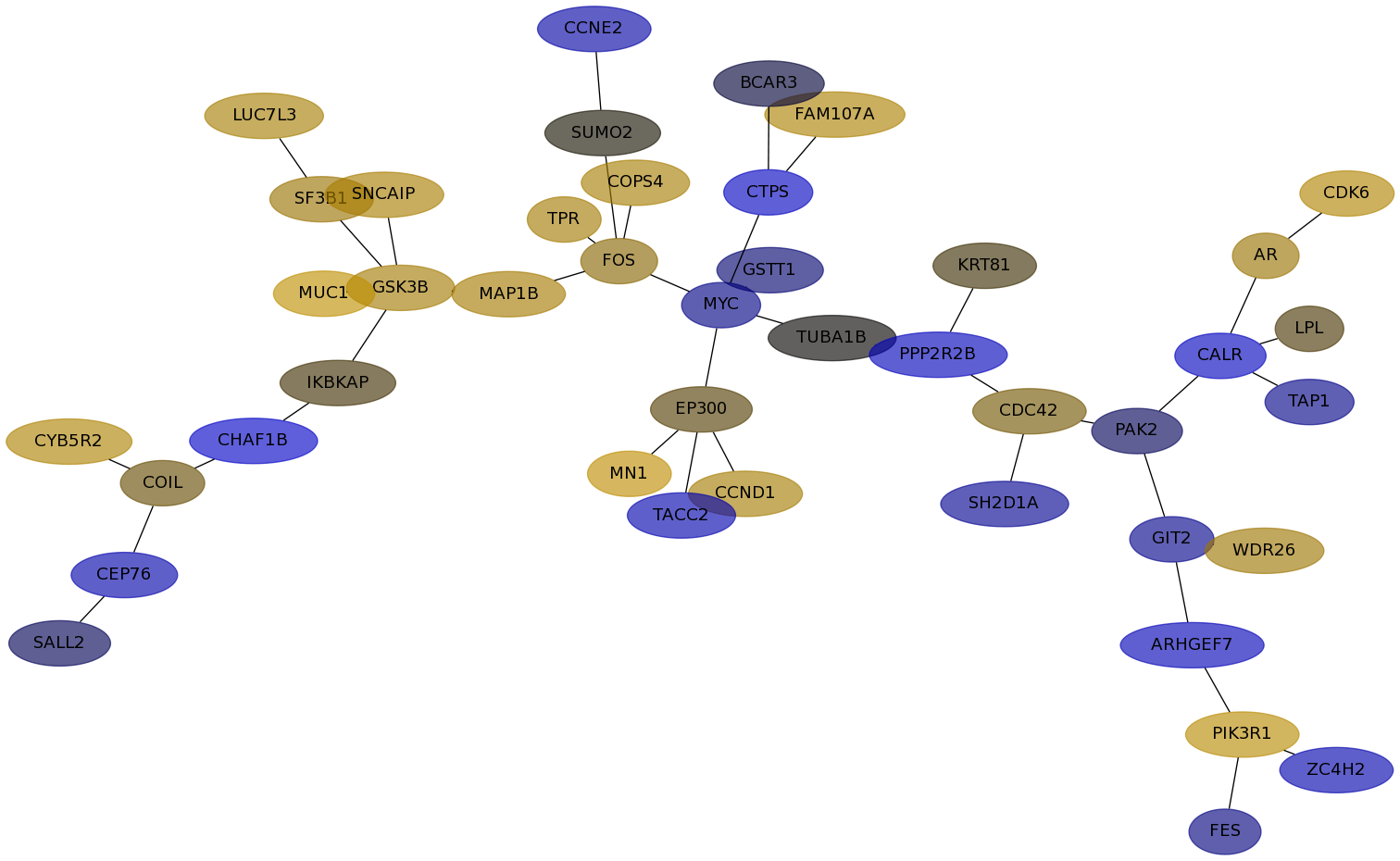
Score for each gene in subnetwork 1169 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| CCNE2 |   | 8 | 84 | 536 | 515 | -0.123 | 0.136 | -0.178 | 0.241 | -0.055 |
|---|
| ARHGEF7 |   | 12 | 55 | 492 | 475 | -0.180 | -0.074 | -0.061 | 0.208 | 0.015 |
|---|
| SF3B1 |   | 15 | 40 | 148 | 144 | 0.091 | -0.154 | 0.126 | -0.073 | -0.204 |
|---|
| SUMO2 |   | 12 | 55 | 148 | 150 | 0.003 | 0.117 | 0.062 | 0.294 | -0.096 |
|---|
| TAP1 |   | 1 | 652 | 584 | 604 | -0.059 | 0.210 | -0.116 | -0.118 | -0.146 |
|---|
| GSTT1 |   | 6 | 120 | 364 | 355 | -0.034 | 0.148 | 0.107 | -0.190 | -0.064 |
|---|
| WDR26 |   | 2 | 391 | 584 | 578 | 0.101 | 0.007 | 0.149 | -0.028 | 0.110 |
|---|
| IKBKAP |   | 5 | 148 | 364 | 358 | 0.010 | 0.089 | 0.184 | 0.142 | -0.017 |
|---|
| KRT81 |   | 2 | 391 | 182 | 195 | 0.009 | 0.108 | -0.105 | 0.252 | 0.086 |
|---|
| GIT2 |   | 3 | 265 | 584 | 573 | -0.065 | -0.191 | 0.362 | -0.164 | 0.054 |
|---|
| SALL2 |   | 1 | 652 | 584 | 604 | -0.018 | -0.058 | 0.015 | 0.209 | -0.058 |
|---|
| CALR |   | 10 | 67 | 117 | 121 | -0.216 | -0.105 | 0.002 | -0.061 | -0.075 |
|---|
| CCND1 |   | 12 | 55 | 148 | 150 | 0.122 | -0.258 | 0.074 | 0.117 | 0.003 |
|---|
| SH2D1A |   | 2 | 391 | 584 | 578 | -0.074 | -0.078 | -0.261 | -0.082 | -0.033 |
|---|
| COPS4 |   | 1 | 652 | 584 | 604 | 0.126 | 0.158 | 0.008 | 0.107 | 0.058 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| TACC2 |   | 9 | 77 | 148 | 154 | -0.151 | -0.073 | 0.122 | 0.181 | 0.180 |
|---|
| MYC |   | 24 | 21 | 148 | 137 | -0.056 | -0.101 | -0.169 | 0.188 | -0.134 |
|---|
| PAK2 |   | 20 | 24 | 1 | 11 | -0.019 | 0.014 | 0.161 | 0.081 | 0.062 |
|---|
| CTPS |   | 2 | 391 | 584 | 578 | -0.227 | 0.060 | -0.110 | 0.026 | -0.041 |
|---|
| FAM107A |   | 2 | 391 | 398 | 411 | 0.142 | -0.102 | -0.043 | 0.144 | -0.014 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| CDK6 |   | 25 | 19 | 1 | 10 | 0.156 | -0.229 | 0.107 | -0.028 | 0.104 |
|---|
| FES |   | 1 | 652 | 584 | 604 | -0.048 | -0.016 | -0.102 | 0.229 | -0.009 |
|---|
| MN1 |   | 6 | 120 | 148 | 156 | 0.213 | -0.166 | -0.047 | 0.150 | 0.011 |
|---|
| CHAF1B |   | 2 | 391 | 584 | 578 | -0.264 | 0.015 | -0.193 | 0.171 | -0.041 |
|---|
| ZC4H2 |   | 16 | 34 | 339 | 332 | -0.145 | 0.065 | 0.061 | 0.304 | 0.059 |
|---|
| CYB5R2 |   | 2 | 391 | 584 | 578 | 0.147 | 0.164 | 0.230 | 0.006 | 0.016 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| CEP76 |   | 1 | 652 | 584 | 604 | -0.140 | 0.055 | -0.234 | 0.270 | 0.020 |
|---|
| FOS |   | 20 | 24 | 148 | 138 | 0.060 | -0.137 | 0.129 | 0.185 | 0.234 |
|---|
| MAP1B |   | 13 | 47 | 148 | 148 | 0.118 | -0.123 | 0.263 | -0.107 | 0.100 |
|---|
| TUBA1B |   | 7 | 100 | 182 | 180 | 0.001 | 0.116 | 0.094 | 0.249 | 0.128 |
|---|
| BCAR3 |   | 2 | 391 | 584 | 578 | -0.009 | 0.271 | 0.123 | 0.259 | 0.066 |
|---|
| TPR |   | 10 | 67 | 148 | 152 | 0.118 | -0.130 | 0.120 | -0.036 | 0.156 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| LUC7L3 |   | 13 | 47 | 148 | 148 | 0.123 | 0.043 | 0.237 | 0.175 | 0.074 |
|---|
| AR |   | 9 | 77 | 283 | 277 | 0.089 | -0.163 | 0.081 | -0.035 | -0.030 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| LPL |   | 4 | 198 | 584 | 571 | 0.013 | -0.102 | -0.062 | 0.292 | -0.105 |
|---|
| SNCAIP |   | 4 | 198 | 283 | 289 | 0.127 | -0.091 | 0.013 | 0.092 | 0.059 |
|---|
| COIL |   | 3 | 265 | 584 | 573 | 0.027 | 0.151 | 0.144 | 0.071 | 0.044 |
|---|
GO Enrichment output for subnetwork 1169 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 1.025E-06 | 2.357E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.025E-06 | 1.179E-03 |
|---|
| macrophage differentiation | GO:0030225 |  | 2.043E-06 | 1.566E-03 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 3.563E-06 | 2.049E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 3.563E-06 | 1.639E-03 |
|---|
| nucleus localization | GO:0051647 |  | 8.492E-06 | 3.255E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 1.209E-05 | 3.972E-03 |
|---|
| protein sumoylation | GO:0016925 |  | 1.209E-05 | 3.476E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 1.657E-05 | 4.234E-03 |
|---|
| microspike assembly | GO:0030035 |  | 2.201E-05 | 5.063E-03 |
|---|
| regulation of erythrocyte differentiation | GO:0045646 |  | 3.617E-05 | 7.563E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 7.831E-07 | 1.913E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 7.831E-07 | 9.566E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.182E-06 | 1.777E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 2.182E-06 | 1.332E-03 |
|---|
| macrophage differentiation | GO:0030225 |  | 3.264E-06 | 1.595E-03 |
|---|
| protein sumoylation | GO:0016925 |  | 4.652E-06 | 1.894E-03 |
|---|
| nucleus localization | GO:0051647 |  | 6.38E-06 | 2.227E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 8.485E-06 | 2.591E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 2.59E-05 | 7.03E-03 |
|---|
| microspike assembly | GO:0030035 |  | 3.1E-05 | 7.574E-03 |
|---|
| myeloid leukocyte differentiation | GO:0002573 |  | 8.586E-05 | 0.01906938 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 1.076E-06 | 2.588E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.076E-06 | 1.294E-03 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.877E-06 | 1.505E-03 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.877E-06 | 1.129E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 2.995E-06 | 1.441E-03 |
|---|
| protein sumoylation | GO:0016925 |  | 6.382E-06 | 2.559E-03 |
|---|
| nucleus localization | GO:0051647 |  | 8.751E-06 | 3.008E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 1.164E-05 | 3.5E-03 |
|---|
| regulation of erythrocyte differentiation | GO:0045646 |  | 1.915E-05 | 5.118E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 2.929E-05 | 7.047E-03 |
|---|
| microspike assembly | GO:0030035 |  | 3.547E-05 | 7.758E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 1.025E-06 | 2.357E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.025E-06 | 1.179E-03 |
|---|
| macrophage differentiation | GO:0030225 |  | 2.043E-06 | 1.566E-03 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 3.563E-06 | 2.049E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 3.563E-06 | 1.639E-03 |
|---|
| nucleus localization | GO:0051647 |  | 8.492E-06 | 3.255E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 1.209E-05 | 3.972E-03 |
|---|
| protein sumoylation | GO:0016925 |  | 1.209E-05 | 3.476E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 1.657E-05 | 4.234E-03 |
|---|
| microspike assembly | GO:0030035 |  | 2.201E-05 | 5.063E-03 |
|---|
| regulation of erythrocyte differentiation | GO:0045646 |  | 3.617E-05 | 7.563E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 1.025E-06 | 2.357E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.025E-06 | 1.179E-03 |
|---|
| macrophage differentiation | GO:0030225 |  | 2.043E-06 | 1.566E-03 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 3.563E-06 | 2.049E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 3.563E-06 | 1.639E-03 |
|---|
| nucleus localization | GO:0051647 |  | 8.492E-06 | 3.255E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 1.209E-05 | 3.972E-03 |
|---|
| protein sumoylation | GO:0016925 |  | 1.209E-05 | 3.476E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 1.657E-05 | 4.234E-03 |
|---|
| microspike assembly | GO:0030035 |  | 2.201E-05 | 5.063E-03 |
|---|
| regulation of erythrocyte differentiation | GO:0045646 |  | 3.617E-05 | 7.563E-03 |
|---|


