Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1137
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3338 | 1.479e-03 | 2.600e-05 | 1.610e-01 | 6.191e-09 |
|---|
| IPC | 0.3386 | 3.524e-01 | 1.816e-01 | 9.364e-01 | 5.991e-02 |
|---|
| Loi | 0.4749 | 5.900e-05 | 0.000e+00 | 5.416e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5978 | 1.000e-06 | 0.000e+00 | 1.174e-01 | 0.000e+00 |
|---|
| Wang | 0.2576 | 7.549e-03 | 6.580e-02 | 2.286e-01 | 1.135e-04 |
|---|
Expression data for subnetwork 1137 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
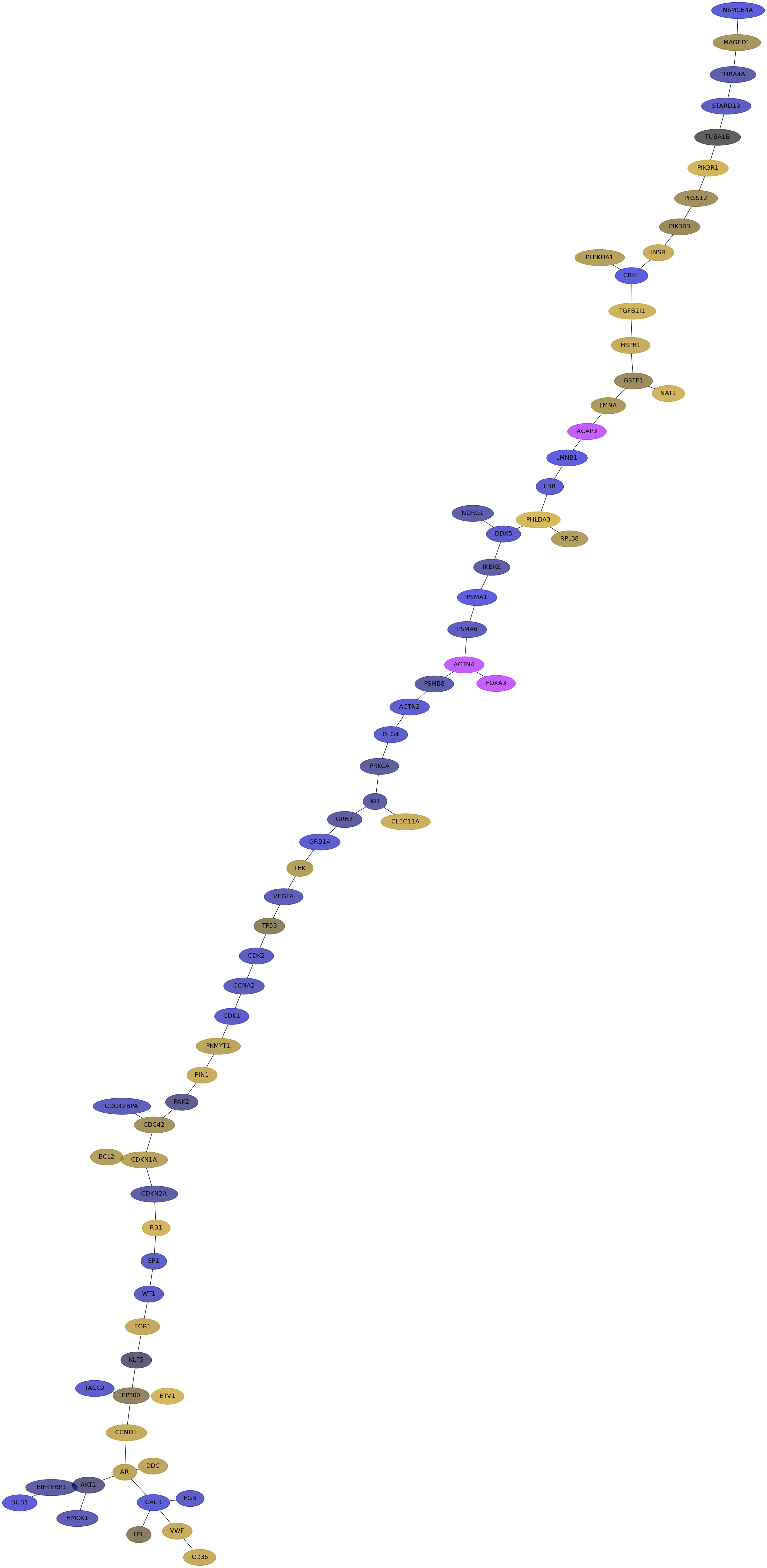
Score for each gene in subnetwork 1137 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| TEK |   | 7 | 100 | 148 | 155 | 0.063 | -0.149 | -0.049 | 0.174 | 0.113 |
|---|
| PRSS12 |   | 1 | 652 | 977 | 985 | 0.032 | -0.069 | 0.006 | -0.049 | -0.189 |
|---|
| NDRG1 |   | 3 | 265 | 756 | 748 | -0.049 | 0.222 | 0.143 | -0.040 | 0.242 |
|---|
| MAGED1 |   | 6 | 120 | 715 | 695 | 0.040 | -0.141 | 0.284 | -0.152 | 0.045 |
|---|
| TP53 |   | 28 | 15 | 1 | 9 | 0.015 | -0.167 | 0.148 | -0.027 | 0.147 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| PRKCA |   | 5 | 148 | 339 | 337 | -0.029 | -0.016 | -0.088 | 0.174 | 0.005 |
|---|
| HMOX1 |   | 2 | 391 | 977 | 968 | -0.063 | 0.109 | -0.120 | -0.110 | 0.152 |
|---|
| CRKL |   | 7 | 100 | 326 | 317 | -0.271 | -0.023 | 0.056 | 0.271 | -0.008 |
|---|
| CCND1 |   | 12 | 55 | 148 | 150 | 0.122 | -0.258 | 0.074 | 0.117 | 0.003 |
|---|
| PSMA6 |   | 2 | 391 | 977 | 968 | -0.104 | 0.280 | -0.118 | -0.018 | -0.091 |
|---|
| PSMB8 |   | 4 | 198 | 710 | 696 | -0.039 | 0.254 | -0.071 | -0.088 | -0.139 |
|---|
| CCNA2 |   | 15 | 40 | 148 | 144 | -0.097 | 0.176 | -0.053 | 0.188 | 0.246 |
|---|
| LMNB1 |   | 2 | 391 | 412 | 423 | -0.286 | -0.158 | -0.177 | 0.010 | 0.156 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| TACC2 |   | 9 | 77 | 148 | 154 | -0.151 | -0.073 | 0.122 | 0.181 | 0.180 |
|---|
| PIN1 |   | 17 | 30 | 1 | 13 | 0.135 | 0.194 | 0.040 | -0.040 | -0.048 |
|---|
| PIK3R3 |   | 1 | 652 | 977 | 985 | 0.023 | -0.048 | 0.212 | -0.036 | -0.075 |
|---|
| ETV1 |   | 11 | 59 | 746 | 724 | 0.214 | 0.046 | 0.054 | -0.065 | 0.141 |
|---|
| KIT |   | 13 | 47 | 339 | 335 | -0.034 | -0.130 | -0.091 | 0.235 | -0.059 |
|---|
| TUBA4A |   | 3 | 265 | 715 | 705 | -0.041 | 0.157 | 0.136 | -0.010 | 0.156 |
|---|
| AKT1 |   | 13 | 47 | 444 | 430 | -0.012 | 0.045 | 0.126 | -0.099 | -0.168 |
|---|
| RPL38 |   | 3 | 265 | 859 | 843 | 0.065 | 0.222 | 0.151 | 0.241 | -0.171 |
|---|
| DLG4 |   | 1 | 652 | 977 | 985 | -0.128 | 0.059 | -0.113 | 0.151 | 0.123 |
|---|
| PKMYT1 |   | 5 | 148 | 620 | 603 | 0.085 | 0.175 | -0.143 | 0.046 | -0.070 |
|---|
| PLEKHA1 |   | 2 | 391 | 746 | 751 | 0.068 | 0.208 | -0.052 | 0.166 | 0.179 |
|---|
| VEGFA |   | 28 | 15 | 57 | 49 | -0.074 | 0.090 | 0.200 | 0.030 | 0.148 |
|---|
| CD36 |   | 1 | 652 | 977 | 985 | 0.119 | 0.146 | 0.017 | 0.129 | 0.124 |
|---|
| CDK1 |   | 16 | 34 | 206 | 194 | -0.138 | 0.265 | -0.195 | 0.106 | 0.031 |
|---|
| TUBA1B |   | 7 | 100 | 182 | 180 | 0.001 | 0.116 | 0.094 | 0.249 | 0.128 |
|---|
| EIF4EBP1 |   | 3 | 265 | 444 | 447 | -0.030 | 0.086 | -0.201 | -0.147 | 0.216 |
|---|
| ACTN4 |   | 6 | 120 | 859 | 834 | undef | 0.035 | 0.180 | undef | undef |
|---|
| KLF5 |   | 25 | 19 | 57 | 50 | -0.005 | 0.077 | 0.117 | -0.007 | -0.006 |
|---|
| LPL |   | 4 | 198 | 584 | 571 | 0.013 | -0.102 | -0.062 | 0.292 | -0.105 |
|---|
| STARD13 |   | 2 | 391 | 859 | 860 | -0.108 | -0.083 | 0.006 | 0.126 | 0.028 |
|---|
| HSPB1 |   | 4 | 198 | 148 | 162 | 0.121 | 0.053 | 0.169 | 0.017 | 0.161 |
|---|
| TGFB1I1 |   | 16 | 34 | 326 | 315 | 0.176 | -0.105 | 0.229 | 0.143 | 0.298 |
|---|
| NSMCE4A |   | 3 | 265 | 715 | 705 | -0.228 | 0.177 | -0.116 | 0.290 | -0.091 |
|---|
| EGR1 |   | 11 | 59 | 283 | 274 | 0.118 | -0.184 | 0.259 | 0.130 | 0.122 |
|---|
| GRB14 |   | 4 | 198 | 977 | 965 | -0.189 | 0.113 | -0.129 | 0.143 | 0.138 |
|---|
| FOXA3 |   | 6 | 120 | 657 | 631 | undef | 0.192 | 0.289 | undef | undef |
|---|
| ACTN2 |   | 2 | 391 | 977 | 968 | -0.198 | 0.139 | -0.106 | 0.155 | 0.127 |
|---|
| GSTP1 |   | 1 | 652 | 977 | 985 | 0.025 | 0.222 | -0.166 | 0.201 | -0.136 |
|---|
| LBR |   | 1 | 652 | 977 | 985 | -0.171 | 0.211 | -0.170 | 0.125 | 0.094 |
|---|
| LMNA |   | 1 | 652 | 977 | 985 | 0.049 | 0.208 | 0.223 | -0.017 | 0.069 |
|---|
| CALR |   | 10 | 67 | 117 | 121 | -0.216 | -0.105 | 0.002 | -0.061 | -0.075 |
|---|
| RB1 |   | 26 | 18 | 271 | 263 | 0.200 | 0.013 | -0.051 | 0.057 | -0.082 |
|---|
| ACAP3 |   | 1 | 652 | 977 | 985 | undef | 0.064 | 0.180 | undef | undef |
|---|
| PAK2 |   | 20 | 24 | 1 | 11 | -0.019 | 0.014 | 0.161 | 0.081 | 0.062 |
|---|
| WT1 |   | 8 | 84 | 283 | 278 | -0.106 | 0.254 | -0.039 | 0.036 | 0.172 |
|---|
| BUB1 |   | 1 | 652 | 977 | 985 | -0.236 | 0.246 | -0.172 | 0.013 | 0.170 |
|---|
| BCL2 |   | 11 | 59 | 364 | 350 | 0.067 | -0.077 | -0.035 | 0.128 | 0.043 |
|---|
| DDC |   | 4 | 198 | 715 | 700 | 0.093 | -0.121 | 0.157 | -0.078 | 0.114 |
|---|
| SP1 |   | 16 | 34 | 283 | 272 | -0.129 | 0.222 | -0.254 | 0.239 | 0.082 |
|---|
| CLEC11A |   | 5 | 148 | 339 | 337 | 0.146 | 0.004 | 0.353 | 0.005 | 0.272 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| PSMA1 |   | 1 | 652 | 977 | 985 | -0.283 | 0.129 | -0.118 | 0.125 | -0.082 |
|---|
| CDC42BPA |   | 2 | 391 | 756 | 756 | -0.082 | -0.102 | 0.098 | 0.059 | 0.260 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| IKBKE |   | 8 | 84 | 756 | 726 | -0.033 | 0.227 | -0.041 | -0.095 | 0.079 |
|---|
| NAT1 |   | 1 | 652 | 977 | 985 | 0.182 | 0.175 | 0.043 | 0.032 | 0.011 |
|---|
| GRB7 |   | 13 | 47 | 57 | 51 | -0.028 | 0.021 | 0.115 | -0.072 | 0.129 |
|---|
| AR |   | 9 | 77 | 283 | 277 | 0.089 | -0.163 | 0.081 | -0.035 | -0.030 |
|---|
| VWF |   | 4 | 198 | 57 | 68 | 0.126 | -0.091 | 0.072 | 0.152 | 0.100 |
|---|
| FGB |   | 3 | 265 | 910 | 895 | -0.100 | 0.136 | 0.231 | -0.035 | -0.016 |
|---|
| DDX5 |   | 1 | 652 | 977 | 985 | -0.144 | -0.112 | 0.153 | 0.021 | -0.132 |
|---|
| CDK2 |   | 16 | 34 | 129 | 124 | -0.104 | 0.196 | -0.096 | 0.109 | 0.362 |
|---|
| CDKN2A |   | 10 | 67 | 364 | 351 | -0.037 | 0.263 | -0.113 | 0.111 | -0.105 |
|---|
| PHLDA3 |   | 1 | 652 | 977 | 985 | 0.228 | -0.017 | 0.204 | -0.102 | -0.069 |
|---|
GO Enrichment output for subnetwork 1137 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 1.497E-08 | 3.443E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.907E-08 | 3.342E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.036E-07 | 7.941E-05 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.036E-07 | 5.956E-05 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 1.734E-07 | 7.975E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 2.06E-07 | 7.895E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 2.186E-07 | 7.182E-05 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.471E-07 | 7.103E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.605E-07 | 6.658E-05 |
|---|
| interphase | GO:0051325 |  | 3.117E-07 | 7.169E-05 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 3.519E-07 | 7.357E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.879E-09 | 4.59E-06 |
|---|
| nuclear migration | GO:0007097 |  | 6.668E-09 | 8.145E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.007E-08 | 1.634E-05 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 2.972E-08 | 1.815E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 3.089E-08 | 1.509E-05 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 3.089E-08 | 1.258E-05 |
|---|
| negative regulation of cell size | GO:0045792 |  | 3.463E-08 | 1.208E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 3.463E-08 | 1.057E-05 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 3.832E-08 | 1.04E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 5.54E-08 | 1.353E-05 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 5.664E-08 | 1.258E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.916E-09 | 1.183E-05 |
|---|
| nuclear migration | GO:0007097 |  | 1.17E-08 | 1.408E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.719E-08 | 2.181E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.589E-08 | 2.76E-05 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 5.416E-08 | 2.606E-05 |
|---|
| interphase | GO:0051325 |  | 5.861E-08 | 2.35E-05 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 5.96E-08 | 2.049E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 7.271E-08 | 2.187E-05 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 8.069E-08 | 2.157E-05 |
|---|
| negative regulation of cell size | GO:0045792 |  | 8.94E-08 | 2.151E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 9.708E-08 | 2.123E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 1.497E-08 | 3.443E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.907E-08 | 3.342E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.036E-07 | 7.941E-05 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.036E-07 | 5.956E-05 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 1.734E-07 | 7.975E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 2.06E-07 | 7.895E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 2.186E-07 | 7.182E-05 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.471E-07 | 7.103E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.605E-07 | 6.658E-05 |
|---|
| interphase | GO:0051325 |  | 3.117E-07 | 7.169E-05 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 3.519E-07 | 7.357E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 1.497E-08 | 3.443E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.907E-08 | 3.342E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.036E-07 | 7.941E-05 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.036E-07 | 5.956E-05 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 1.734E-07 | 7.975E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 2.06E-07 | 7.895E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 2.186E-07 | 7.182E-05 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.471E-07 | 7.103E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.605E-07 | 6.658E-05 |
|---|
| interphase | GO:0051325 |  | 3.117E-07 | 7.169E-05 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 3.519E-07 | 7.357E-05 |
|---|


