Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1135
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3339 | 1.468e-03 | 2.500e-05 | 1.605e-01 | 5.890e-09 |
|---|
| IPC | 0.3383 | 3.529e-01 | 1.820e-01 | 9.365e-01 | 6.016e-02 |
|---|
| Loi | 0.4747 | 6.000e-05 | 0.000e+00 | 5.430e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5977 | 1.000e-06 | 0.000e+00 | 1.175e-01 | 0.000e+00 |
|---|
| Wang | 0.2575 | 7.576e-03 | 6.595e-02 | 2.290e-01 | 1.144e-04 |
|---|
Expression data for subnetwork 1135 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
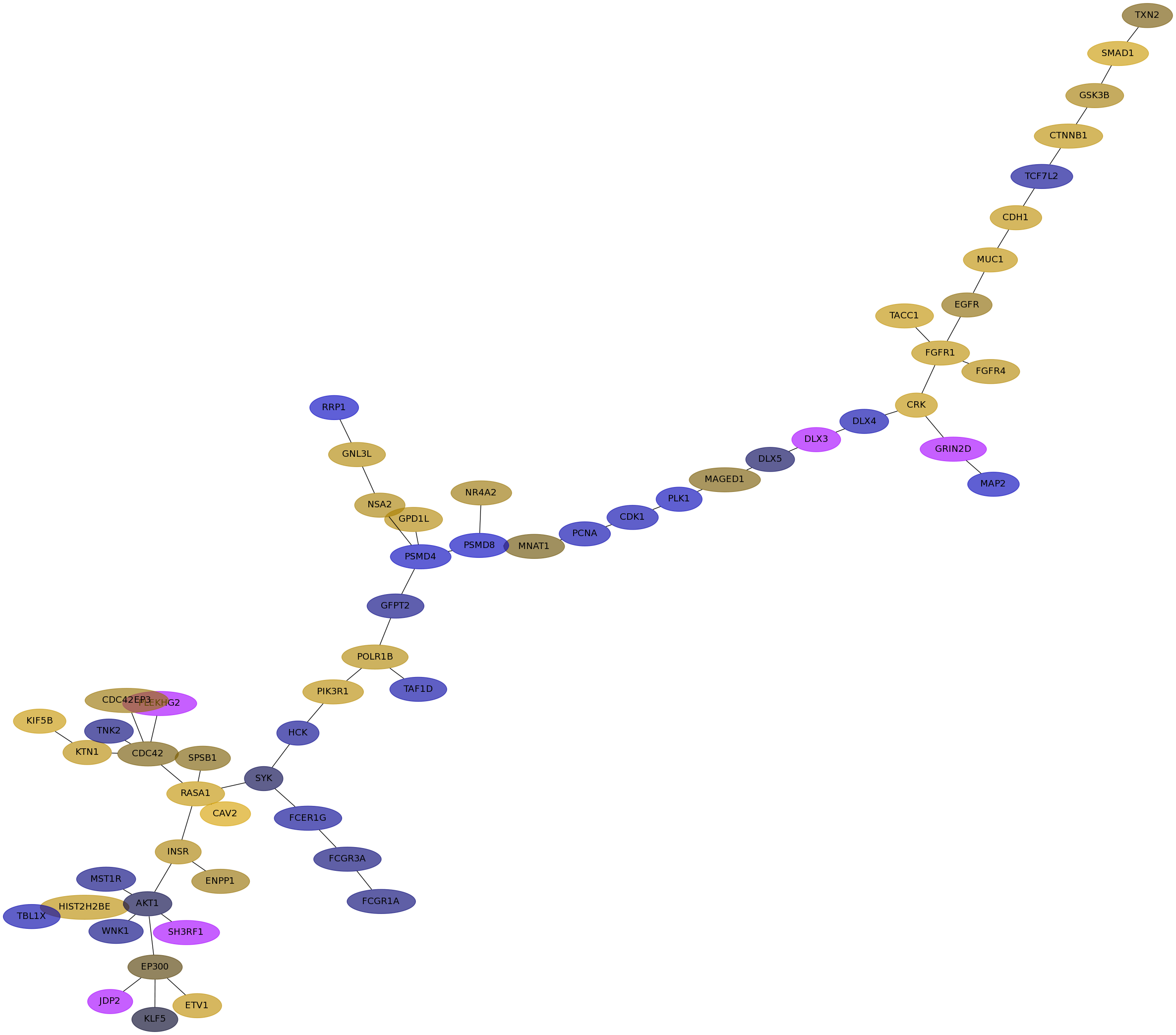
Score for each gene in subnetwork 1135 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| CDC42EP3 |   | 2 | 391 | 1008 | 999 | 0.089 | -0.159 | 0.338 | -0.073 | 0.220 |
|---|
| GFPT2 |   | 1 | 652 | 1008 | 1014 | -0.050 | 0.004 | 0.096 | -0.142 | 0.176 |
|---|
| FCER1G |   | 1 | 652 | 1008 | 1014 | -0.074 | 0.049 | -0.280 | -0.023 | 0.017 |
|---|
| JDP2 |   | 1 | 652 | 1008 | 1014 | undef | 0.062 | 0.045 | undef | undef |
|---|
| MAGED1 |   | 6 | 120 | 715 | 695 | 0.040 | -0.141 | 0.284 | -0.152 | 0.045 |
|---|
| HIST2H2BE |   | 5 | 148 | 492 | 498 | 0.196 | -0.086 | 0.123 | -0.180 | -0.104 |
|---|
| SPSB1 |   | 7 | 100 | 492 | 495 | 0.044 | -0.124 | 0.088 | -0.152 | 0.169 |
|---|
| KTN1 |   | 1 | 652 | 1008 | 1014 | 0.183 | 0.146 | 0.243 | 0.082 | 0.208 |
|---|
| KIF5B |   | 1 | 652 | 1008 | 1014 | 0.266 | 0.003 | 0.010 | -0.104 | -0.149 |
|---|
| WNK1 |   | 1 | 652 | 1008 | 1014 | -0.050 | -0.184 | 0.094 | -0.007 | 0.051 |
|---|
| PCNA |   | 3 | 265 | 57 | 71 | -0.138 | 0.243 | -0.158 | 0.027 | 0.003 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| FCGR1A |   | 2 | 391 | 364 | 375 | -0.037 | 0.297 | -0.237 | -0.097 | 0.060 |
|---|
| POLR1B |   | 11 | 59 | 129 | 126 | 0.157 | 0.062 | -0.036 | -0.079 | 0.139 |
|---|
| DLX4 |   | 2 | 391 | 715 | 711 | -0.133 | 0.291 | 0.057 | -0.004 | -0.059 |
|---|
| GPD1L |   | 1 | 652 | 1008 | 1014 | 0.161 | -0.130 | -0.025 | -0.055 | -0.162 |
|---|
| ETV1 |   | 11 | 59 | 746 | 724 | 0.214 | 0.046 | 0.054 | -0.065 | 0.141 |
|---|
| SYK |   | 1 | 652 | 1008 | 1014 | -0.014 | 0.106 | -0.251 | -0.134 | 0.163 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| PLK1 |   | 5 | 148 | 444 | 440 | -0.179 | 0.142 | -0.143 | 0.094 | -0.163 |
|---|
| TNK2 |   | 1 | 652 | 1008 | 1014 | -0.041 | 0.077 | 0.149 | 0.079 | 0.053 |
|---|
| RRP1 |   | 2 | 391 | 1008 | 999 | -0.224 | 0.154 | -0.104 | 0.095 | -0.135 |
|---|
| AKT1 |   | 13 | 47 | 444 | 430 | -0.012 | 0.045 | 0.126 | -0.099 | -0.168 |
|---|
| TACC1 |   | 5 | 148 | 326 | 330 | 0.229 | -0.051 | 0.020 | -0.022 | 0.126 |
|---|
| TAF1D |   | 5 | 148 | 57 | 63 | -0.110 | 0.141 | 0.113 | 0.041 | 0.059 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| DLX3 |   | 1 | 652 | 1008 | 1014 | undef | 0.144 | -0.056 | undef | undef |
|---|
| PSMD4 |   | 1 | 652 | 1008 | 1014 | -0.202 | 0.285 | -0.243 | -0.043 | -0.045 |
|---|
| HCK |   | 3 | 265 | 598 | 596 | -0.066 | -0.112 | -0.228 | -0.124 | -0.072 |
|---|
| CDK1 |   | 16 | 34 | 206 | 194 | -0.138 | 0.265 | -0.195 | 0.106 | 0.031 |
|---|
| FCGR3A |   | 1 | 652 | 1008 | 1014 | -0.035 | 0.174 | -0.195 | -0.072 | 0.090 |
|---|
| TCF7L2 |   | 17 | 30 | 271 | 264 | -0.071 | -0.021 | 0.143 | 0.280 | 0.101 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| RASA1 |   | 9 | 77 | 492 | 494 | 0.237 | -0.079 | 0.146 | 0.127 | 0.149 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| MNAT1 |   | 2 | 391 | 57 | 80 | 0.029 | 0.194 | 0.055 | 0.224 | 0.236 |
|---|
| NSA2 |   | 1 | 652 | 1008 | 1014 | 0.131 | 0.103 | 0.097 | 0.132 | 0.006 |
|---|
| ENPP1 |   | 3 | 265 | 778 | 770 | 0.082 | 0.088 | 0.129 | -0.026 | 0.039 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| CRK |   | 27 | 17 | 206 | 193 | 0.232 | -0.116 | 0.106 | 0.090 | 0.017 |
|---|
| MST1R |   | 1 | 652 | 1008 | 1014 | -0.046 | 0.124 | 0.083 | -0.212 | 0.026 |
|---|
| MAP2 |   | 1 | 652 | 1008 | 1014 | -0.194 | 0.044 | 0.045 | 0.014 | -0.057 |
|---|
| SH3RF1 |   | 14 | 42 | 117 | 120 | undef | 0.144 | 0.238 | undef | undef |
|---|
| PSMD8 |   | 1 | 652 | 1008 | 1014 | -0.221 | 0.052 | -0.027 | -0.080 | 0.027 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| GRIN2D |   | 18 | 26 | 33 | 32 | undef | 0.058 | 0.155 | 0.000 | undef |
|---|
| NR4A2 |   | 1 | 652 | 1008 | 1014 | 0.089 | 0.002 | -0.053 | -0.105 | 0.015 |
|---|
| KLF5 |   | 25 | 19 | 57 | 50 | -0.005 | 0.077 | 0.117 | -0.007 | -0.006 |
|---|
| GNL3L |   | 3 | 265 | 994 | 971 | 0.158 | 0.140 | 0.020 | -0.264 | -0.031 |
|---|
| DLX5 |   | 2 | 391 | 1008 | 999 | -0.019 | -0.026 | 0.044 | -0.151 | 0.140 |
|---|
| TBL1X |   | 2 | 391 | 1008 | 999 | -0.131 | -0.044 | 0.017 | -0.192 | -0.009 |
|---|
| SMAD1 |   | 24 | 21 | 33 | 30 | 0.287 | 0.049 | 0.159 | -0.062 | 0.048 |
|---|
| TXN2 |   | 4 | 198 | 33 | 41 | 0.035 | -0.112 | -0.009 | -0.044 | -0.139 |
|---|
| PLEKHG2 |   | 1 | 652 | 1008 | 1014 | undef | -0.112 | 0.179 | undef | undef |
|---|
GO Enrichment output for subnetwork 1135 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 8.443E-08 | 1.942E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 8.637E-08 | 9.933E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.025E-07 | 7.856E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.205E-07 | 6.928E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 1.205E-07 | 5.542E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 1.831E-07 | 7.018E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 2.235E-07 | 7.343E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 3.27E-07 | 9.402E-05 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.72E-06 | 4.396E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.727E-06 | 3.971E-04 |
|---|
| nuclear migration | GO:0007097 |  | 2.087E-06 | 4.364E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 4.787E-12 | 1.169E-08 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 4.082E-08 | 4.987E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 4.381E-08 | 3.568E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 5.508E-08 | 3.364E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 5.508E-08 | 2.691E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 8.814E-08 | 3.589E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 1.054E-07 | 3.678E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.614E-07 | 4.93E-05 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 9.063E-07 | 2.46E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.012E-06 | 2.473E-04 |
|---|
| regulation of leukocyte degranulation | GO:0043300 |  | 1.049E-06 | 2.329E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 3.216E-12 | 7.738E-09 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 4.62E-08 | 5.557E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 6.304E-08 | 5.056E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 8.499E-08 | 5.112E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 8.499E-08 | 4.09E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 1.358E-07 | 5.447E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 1.623E-07 | 5.58E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 2.483E-07 | 7.469E-05 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.211E-06 | 3.239E-04 |
|---|
| regulation of leukocyte degranulation | GO:0043300 |  | 1.276E-06 | 3.07E-04 |
|---|
| positive regulation of mast cell activation | GO:0033005 |  | 1.276E-06 | 2.791E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 8.443E-08 | 1.942E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 8.637E-08 | 9.933E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.025E-07 | 7.856E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.205E-07 | 6.928E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 1.205E-07 | 5.542E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 1.831E-07 | 7.018E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 2.235E-07 | 7.343E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 3.27E-07 | 9.402E-05 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.72E-06 | 4.396E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.727E-06 | 3.971E-04 |
|---|
| nuclear migration | GO:0007097 |  | 2.087E-06 | 4.364E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 8.443E-08 | 1.942E-04 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 8.637E-08 | 9.933E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.025E-07 | 7.856E-05 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 1.205E-07 | 6.928E-05 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 1.205E-07 | 5.542E-05 |
|---|
| positive regulation of ligase activity | GO:0051351 |  | 1.831E-07 | 7.018E-05 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 2.235E-07 | 7.343E-05 |
|---|
| regulation of ligase activity | GO:0051340 |  | 3.27E-07 | 9.402E-05 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.72E-06 | 4.396E-04 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.727E-06 | 3.971E-04 |
|---|
| nuclear migration | GO:0007097 |  | 2.087E-06 | 4.364E-04 |
|---|


