Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1132
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3332 | 1.519e-03 | 2.700e-05 | 1.629e-01 | 6.679e-09 |
|---|
| IPC | 0.3390 | 3.518e-01 | 1.810e-01 | 9.363e-01 | 5.962e-02 |
|---|
| Loi | 0.4741 | 6.100e-05 | 0.000e+00 | 5.459e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5987 | 1.000e-06 | 0.000e+00 | 1.154e-01 | 0.000e+00 |
|---|
| Wang | 0.2578 | 7.495e-03 | 6.551e-02 | 2.278e-01 | 1.119e-04 |
|---|
Expression data for subnetwork 1132 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
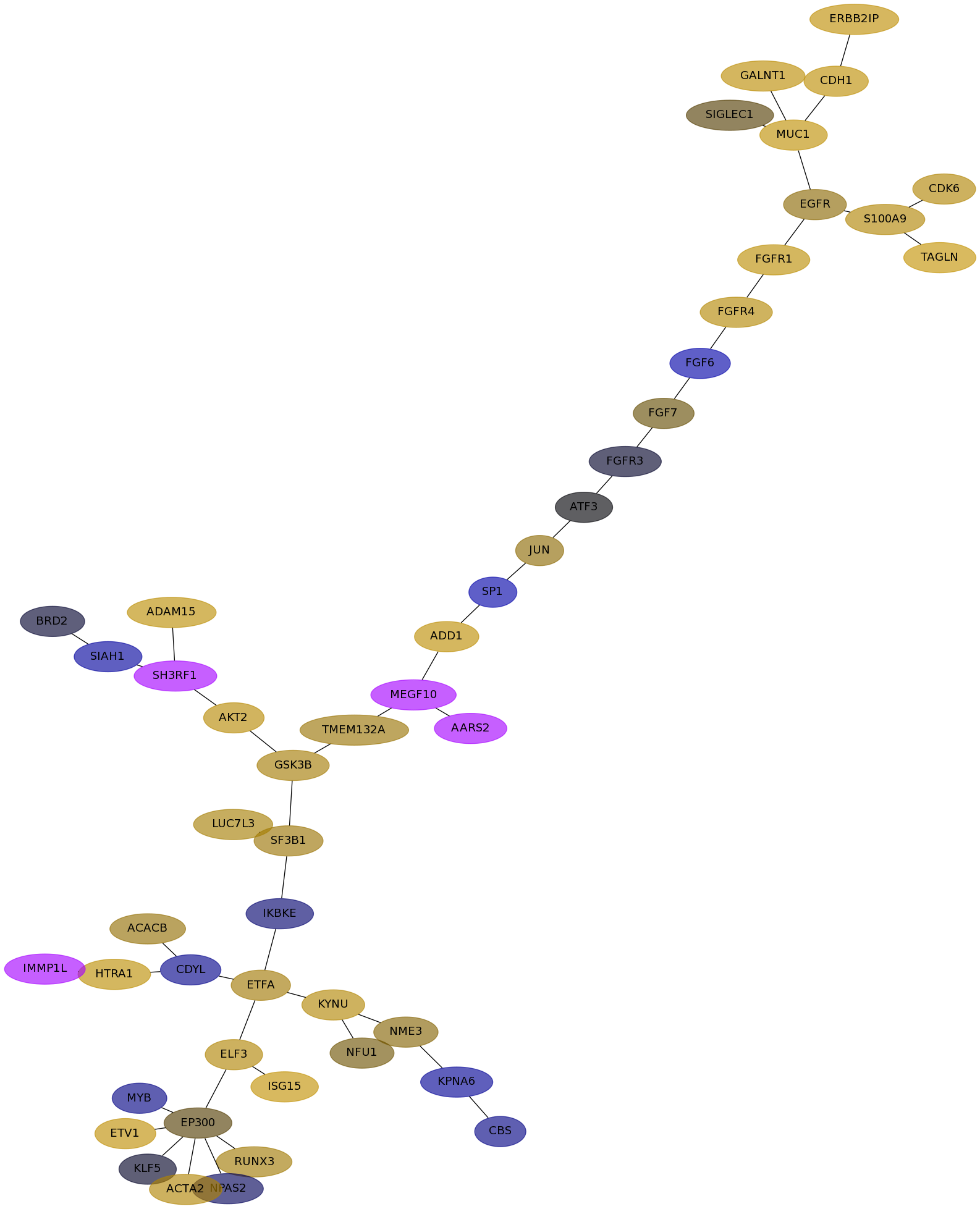
Score for each gene in subnetwork 1132 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| ACACB |   | 1 | 652 | 963 | 973 | 0.078 | -0.069 | 0.025 | 0.127 | -0.006 |
|---|
| ADD1 |   | 2 | 391 | 715 | 711 | 0.209 | -0.085 | 0.116 | 0.035 | -0.098 |
|---|
| NPAS2 |   | 3 | 265 | 364 | 362 | -0.019 | 0.116 | 0.253 | -0.157 | -0.093 |
|---|
| HTRA1 |   | 2 | 391 | 492 | 506 | 0.202 | -0.046 | 0.184 | 0.051 | 0.224 |
|---|
| SF3B1 |   | 15 | 40 | 148 | 144 | 0.091 | -0.154 | 0.126 | -0.073 | -0.204 |
|---|
| AARS2 |   | 1 | 652 | 963 | 973 | undef | 0.014 | 0.053 | undef | undef |
|---|
| CBS |   | 1 | 652 | 963 | 973 | -0.055 | 0.109 | 0.063 | -0.090 | -0.017 |
|---|
| TMEM132A |   | 7 | 100 | 33 | 37 | 0.088 | 0.019 | 0.127 | -0.141 | -0.092 |
|---|
| MEGF10 |   | 1 | 652 | 963 | 973 | undef | 0.047 | -0.066 | undef | undef |
|---|
| ELF3 |   | 2 | 391 | 326 | 344 | 0.154 | 0.178 | -0.022 | -0.052 | -0.076 |
|---|
| JUN |   | 3 | 265 | 283 | 292 | 0.067 | 0.200 | 0.051 | 0.220 | 0.200 |
|---|
| GALNT1 |   | 3 | 265 | 889 | 874 | 0.200 | 0.133 | -0.061 | -0.181 | 0.234 |
|---|
| FGFR3 |   | 5 | 148 | 963 | 938 | -0.006 | 0.160 | 0.071 | 0.018 | -0.111 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| SIAH1 |   | 1 | 652 | 963 | 973 | -0.101 | -0.212 | 0.008 | -0.053 | 0.219 |
|---|
| ETFA |   | 3 | 265 | 756 | 748 | 0.105 | 0.027 | 0.094 | 0.088 | 0.013 |
|---|
| ACTA2 |   | 2 | 391 | 778 | 789 | 0.155 | -0.147 | 0.256 | 0.125 | 0.220 |
|---|
| ADAM15 |   | 13 | 47 | 492 | 474 | 0.205 | -0.048 | -0.078 | -0.208 | -0.048 |
|---|
| NFU1 |   | 2 | 391 | 963 | 966 | 0.032 | 0.280 | 0.023 | 0.085 | -0.257 |
|---|
| S100A9 |   | 18 | 26 | 398 | 382 | 0.154 | 0.117 | 0.043 | -0.110 | -0.045 |
|---|
| ETV1 |   | 11 | 59 | 746 | 724 | 0.214 | 0.046 | 0.054 | -0.065 | 0.141 |
|---|
| CDK6 |   | 25 | 19 | 1 | 10 | 0.156 | -0.229 | 0.107 | -0.028 | 0.104 |
|---|
| MYB |   | 3 | 265 | 148 | 166 | -0.054 | -0.028 | 0.075 | 0.002 | -0.012 |
|---|
| KPNA6 |   | 5 | 148 | 182 | 181 | -0.082 | -0.144 | 0.128 | -0.005 | -0.108 |
|---|
| SP1 |   | 16 | 34 | 283 | 272 | -0.129 | 0.222 | -0.254 | 0.239 | 0.082 |
|---|
| TAGLN |   | 4 | 198 | 823 | 807 | 0.243 | -0.079 | 0.246 | 0.170 | 0.175 |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| FGF6 |   | 2 | 391 | 963 | 966 | -0.131 | 0.125 | -0.051 | -0.159 | -0.050 |
|---|
| AKT2 |   | 3 | 265 | 778 | 770 | 0.179 | 0.110 | -0.118 | -0.003 | -0.092 |
|---|
| CDYL |   | 1 | 652 | 963 | 973 | -0.064 | -0.050 | 0.116 | -0.203 | 0.206 |
|---|
| RUNX3 |   | 8 | 84 | 33 | 35 | 0.109 | 0.071 | -0.097 | 0.083 | -0.074 |
|---|
| ATF3 |   | 1 | 652 | 963 | 973 | -0.001 | 0.225 | -0.021 | 0.000 | -0.039 |
|---|
| ERBB2IP |   | 1 | 652 | 963 | 973 | 0.215 | 0.073 | 0.160 | 0.087 | 0.200 |
|---|
| NME3 |   | 1 | 652 | 963 | 973 | 0.055 | -0.057 | -0.079 | -0.053 | -0.055 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| ISG15 |   | 2 | 391 | 326 | 344 | 0.235 | 0.227 | -0.032 | -0.160 | -0.133 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| IKBKE |   | 8 | 84 | 756 | 726 | -0.033 | 0.227 | -0.041 | -0.095 | 0.079 |
|---|
| BRD2 |   | 3 | 265 | 129 | 139 | -0.006 | 0.016 | 0.348 | 0.105 | 0.093 |
|---|
| SH3RF1 |   | 14 | 42 | 117 | 120 | undef | 0.144 | 0.238 | undef | undef |
|---|
| KYNU |   | 4 | 198 | 606 | 597 | 0.161 | -0.245 | 0.090 | -0.294 | 0.047 |
|---|
| LUC7L3 |   | 13 | 47 | 148 | 148 | 0.123 | 0.043 | 0.237 | 0.175 | 0.074 |
|---|
| FGF7 |   | 8 | 84 | 398 | 384 | 0.026 | -0.094 | 0.237 | 0.070 | 0.083 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| KLF5 |   | 25 | 19 | 57 | 50 | -0.005 | 0.077 | 0.117 | -0.007 | -0.006 |
|---|
| IMMP1L |   | 1 | 652 | 963 | 973 | undef | 0.243 | -0.028 | undef | undef |
|---|
| SIGLEC1 |   | 3 | 265 | 963 | 950 | 0.017 | 0.045 | -0.114 | -0.107 | -0.052 |
|---|
GO Enrichment output for subnetwork 1132 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 4.163E-11 | 9.576E-08 |
|---|
| negative regulation of epithelial cell proliferation | GO:0050680 |  | 5.182E-08 | 5.959E-05 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 7.023E-08 | 5.384E-05 |
|---|
| cell growth | GO:0016049 |  | 2.35E-06 | 1.351E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 3.272E-06 | 1.505E-03 |
|---|
| regulation of protein ubiquitination | GO:0031396 |  | 7.179E-05 | 0.02751814 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 8.8E-05 | 0.02891336 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 1.064E-04 | 0.0305969 |
|---|
| peptidyl-serine modification | GO:0018209 |  | 1.064E-04 | 0.02719724 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 1.272E-04 | 0.02925492 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 1.504E-04 | 0.03145507 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 5.054E-13 | 1.235E-09 |
|---|
| negative regulation of epithelial cell proliferation | GO:0050680 |  | 1.801E-08 | 2.2E-05 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 4.949E-08 | 4.031E-05 |
|---|
| cell growth | GO:0016049 |  | 1.927E-06 | 1.177E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 2.977E-06 | 1.455E-03 |
|---|
| regulation of protein ubiquitination | GO:0031396 |  | 4.625E-05 | 0.01883234 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 9.293E-05 | 0.0324339 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 1.247E-04 | 0.03808792 |
|---|
| peptidyl-serine modification | GO:0018209 |  | 1.247E-04 | 0.03385593 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 1.247E-04 | 0.03047034 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 1.629E-04 | 0.03616926 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.396E-11 | 3.36E-08 |
|---|
| negative regulation of epithelial cell proliferation | GO:0050680 |  | 1.857E-08 | 2.234E-05 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 3.859E-08 | 3.095E-05 |
|---|
| cell growth | GO:0016049 |  | 1.777E-06 | 1.069E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 3.043E-06 | 1.464E-03 |
|---|
| regulation of protein ubiquitination | GO:0031396 |  | 4.726E-05 | 0.01895227 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 9.496E-05 | 0.03263826 |
|---|
| peptidyl-serine modification | GO:0018209 |  | 9.496E-05 | 0.02855847 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 9.496E-05 | 0.02538531 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 1.274E-04 | 0.03066129 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 1.274E-04 | 0.0278739 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 4.163E-11 | 9.576E-08 |
|---|
| negative regulation of epithelial cell proliferation | GO:0050680 |  | 5.182E-08 | 5.959E-05 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 7.023E-08 | 5.384E-05 |
|---|
| cell growth | GO:0016049 |  | 2.35E-06 | 1.351E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 3.272E-06 | 1.505E-03 |
|---|
| regulation of protein ubiquitination | GO:0031396 |  | 7.179E-05 | 0.02751814 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 8.8E-05 | 0.02891336 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 1.064E-04 | 0.0305969 |
|---|
| peptidyl-serine modification | GO:0018209 |  | 1.064E-04 | 0.02719724 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 1.272E-04 | 0.02925492 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 1.504E-04 | 0.03145507 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 4.163E-11 | 9.576E-08 |
|---|
| negative regulation of epithelial cell proliferation | GO:0050680 |  | 5.182E-08 | 5.959E-05 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 7.023E-08 | 5.384E-05 |
|---|
| cell growth | GO:0016049 |  | 2.35E-06 | 1.351E-03 |
|---|
| positive regulation of protein ubiquitination | GO:0031398 |  | 3.272E-06 | 1.505E-03 |
|---|
| regulation of protein ubiquitination | GO:0031396 |  | 7.179E-05 | 0.02751814 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 8.8E-05 | 0.02891336 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 1.064E-04 | 0.0305969 |
|---|
| peptidyl-serine modification | GO:0018209 |  | 1.064E-04 | 0.02719724 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 1.272E-04 | 0.02925492 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 1.504E-04 | 0.03145507 |
|---|


