Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1098
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3349 | 1.406e-03 | 2.300e-05 | 1.575e-01 | 5.094e-09 |
|---|
| IPC | 0.3412 | 3.481e-01 | 1.778e-01 | 9.355e-01 | 5.792e-02 |
|---|
| Loi | 0.4713 | 7.000e-05 | 0.000e+00 | 5.602e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5958 | 1.000e-06 | 0.000e+00 | 1.216e-01 | 0.000e+00 |
|---|
| Wang | 0.2563 | 7.971e-03 | 6.807e-02 | 2.344e-01 | 1.272e-04 |
|---|
Expression data for subnetwork 1098 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
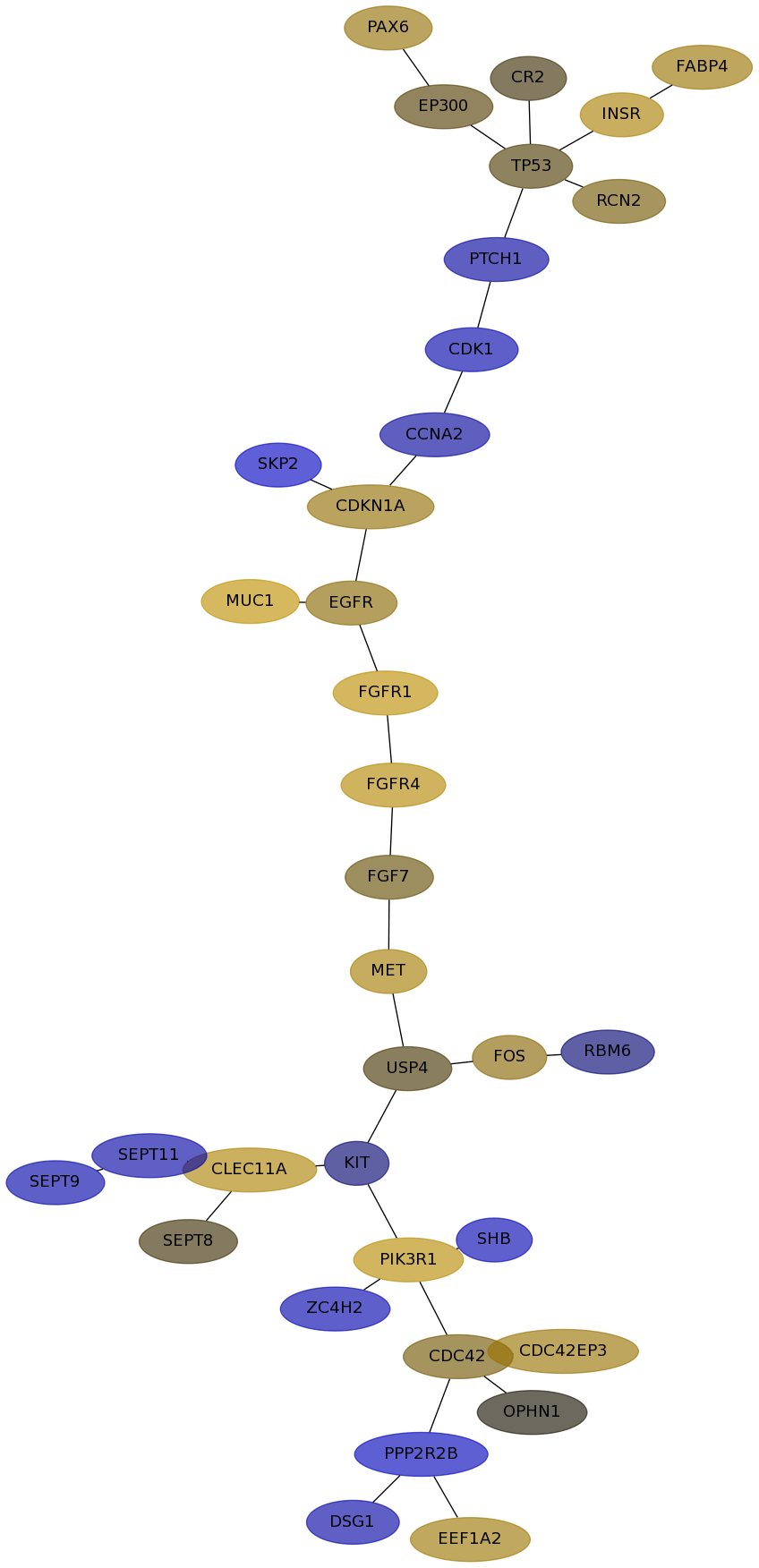
Score for each gene in subnetwork 1098 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| SHB |   | 2 | 391 | 339 | 352 | -0.160 | 0.164 | 0.141 | -0.074 | -0.048 |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| CDC42EP3 |   | 2 | 391 | 1008 | 999 | 0.089 | -0.159 | 0.338 | -0.073 | 0.220 |
|---|
| USP4 |   | 6 | 120 | 823 | 803 | 0.012 | -0.093 | 0.147 | 0.097 | 0.054 |
|---|
| PAX6 |   | 11 | 59 | 283 | 274 | 0.081 | 0.259 | -0.003 | 0.112 | 0.168 |
|---|
| OPHN1 |   | 5 | 148 | 339 | 337 | 0.003 | 0.062 | 0.278 | 0.194 | -0.182 |
|---|
| TP53 |   | 28 | 15 | 1 | 9 | 0.015 | -0.167 | 0.148 | -0.027 | 0.147 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| CCNA2 |   | 15 | 40 | 148 | 144 | -0.097 | 0.176 | -0.053 | 0.188 | 0.246 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| EEF1A2 |   | 9 | 77 | 182 | 179 | 0.098 | 0.032 | 0.220 | -0.119 | 0.102 |
|---|
| RBM6 |   | 10 | 67 | 148 | 152 | -0.036 | -0.082 | 0.265 | -0.004 | 0.111 |
|---|
| SEPT11 |   | 2 | 391 | 1063 | 1057 | -0.123 | -0.075 | -0.054 | 0.193 | 0.048 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| MET |   | 5 | 148 | 536 | 536 | 0.119 | -0.219 | 0.086 | 0.183 | 0.170 |
|---|
| KIT |   | 13 | 47 | 339 | 335 | -0.034 | -0.130 | -0.091 | 0.235 | -0.059 |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| CLEC11A |   | 5 | 148 | 339 | 337 | 0.146 | 0.004 | 0.353 | 0.005 | 0.272 |
|---|
| SEPT9 |   | 2 | 391 | 1063 | 1057 | -0.129 | -0.139 | -0.080 | 0.267 | -0.084 |
|---|
| ZC4H2 |   | 16 | 34 | 339 | 332 | -0.145 | 0.065 | 0.061 | 0.304 | 0.059 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| SKP2 |   | 14 | 42 | 148 | 146 | -0.228 | 0.092 | -0.043 | 0.237 | -0.009 |
|---|
| CR2 |   | 2 | 391 | 847 | 841 | 0.010 | -0.042 | -0.089 | 0.101 | 0.015 |
|---|
| PTCH1 |   | 4 | 198 | 339 | 341 | -0.102 | -0.184 | 0.047 | 0.121 | 0.004 |
|---|
| SEPT8 |   | 2 | 391 | 339 | 352 | 0.010 | 0.209 | 0.148 | 0.097 | 0.169 |
|---|
| FOS |   | 20 | 24 | 148 | 138 | 0.060 | -0.137 | 0.129 | 0.185 | 0.234 |
|---|
| CDK1 |   | 16 | 34 | 206 | 194 | -0.138 | 0.265 | -0.195 | 0.106 | 0.031 |
|---|
| RCN2 |   | 6 | 120 | 756 | 731 | 0.037 | 0.059 | 0.051 | 0.176 | -0.098 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| FGF7 |   | 8 | 84 | 398 | 384 | 0.026 | -0.094 | 0.237 | 0.070 | 0.083 |
|---|
| DSG1 |   | 4 | 198 | 182 | 183 | -0.120 | 0.072 | -0.075 | 0.252 | 0.068 |
|---|
| FABP4 |   | 3 | 265 | 444 | 447 | 0.091 | -0.036 | -0.028 | 0.227 | 0.107 |
|---|
GO Enrichment output for subnetwork 1098 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 6.659E-07 | 1.532E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 6.659E-07 | 7.658E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.328E-06 | 1.018E-03 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.317E-06 | 1.332E-03 |
|---|
| protein heterooligomerization | GO:0051291 |  | 2.317E-06 | 1.066E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 2.317E-06 | 8.881E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.64E-06 | 8.675E-04 |
|---|
| nucleus localization | GO:0051647 |  | 5.528E-06 | 1.589E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 7.873E-06 | 2.012E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 1.079E-05 | 2.483E-03 |
|---|
| microspike assembly | GO:0030035 |  | 1.435E-05 | 3E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.293E-08 | 5.603E-05 |
|---|
| nuclear migration | GO:0007097 |  | 4.978E-07 | 6.08E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 4.978E-07 | 4.053E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.388E-06 | 8.476E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.388E-06 | 6.78E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 2.077E-06 | 8.457E-04 |
|---|
| protein heterooligomerization | GO:0051291 |  | 2.961E-06 | 1.033E-03 |
|---|
| nucleus localization | GO:0051647 |  | 4.063E-06 | 1.241E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 5.405E-06 | 1.467E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.066E-05 | 2.605E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 1.653E-05 | 3.671E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 5.663E-07 | 1.362E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 5.663E-07 | 6.812E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 9.888E-07 | 7.93E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 9.888E-07 | 5.947E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.025E-06 | 4.934E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.578E-06 | 6.33E-04 |
|---|
| protein heterooligomerization | GO:0051291 |  | 2.362E-06 | 8.12E-04 |
|---|
| nucleus localization | GO:0051647 |  | 4.62E-06 | 1.389E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 6.146E-06 | 1.643E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.259E-05 | 3.029E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 1.55E-05 | 3.391E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 6.659E-07 | 1.532E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 6.659E-07 | 7.658E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.328E-06 | 1.018E-03 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.317E-06 | 1.332E-03 |
|---|
| protein heterooligomerization | GO:0051291 |  | 2.317E-06 | 1.066E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 2.317E-06 | 8.881E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.64E-06 | 8.675E-04 |
|---|
| nucleus localization | GO:0051647 |  | 5.528E-06 | 1.589E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 7.873E-06 | 2.012E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 1.079E-05 | 2.483E-03 |
|---|
| microspike assembly | GO:0030035 |  | 1.435E-05 | 3E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 6.659E-07 | 1.532E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 6.659E-07 | 7.658E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.328E-06 | 1.018E-03 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.317E-06 | 1.332E-03 |
|---|
| protein heterooligomerization | GO:0051291 |  | 2.317E-06 | 1.066E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 2.317E-06 | 8.881E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.64E-06 | 8.675E-04 |
|---|
| nucleus localization | GO:0051647 |  | 5.528E-06 | 1.589E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 7.873E-06 | 2.012E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 1.079E-05 | 2.483E-03 |
|---|
| microspike assembly | GO:0030035 |  | 1.435E-05 | 3E-03 |
|---|


