Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6937
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7488 | 3.845e-03 | 6.420e-04 | 7.830e-01 | 1.933e-06 |
|---|
| Loi | 0.2307 | 7.963e-02 | 1.119e-02 | 4.267e-01 | 3.803e-04 |
|---|
| Schmidt | 0.6764 | 0.000e+00 | 0.000e+00 | 4.182e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7912 | 0.000e+00 | 0.000e+00 | 3.417e-03 | 0.000e+00 |
|---|
| Wang | 0.2640 | 2.372e-03 | 4.095e-02 | 3.827e-01 | 3.716e-05 |
|---|
Expression data for subnetwork 6937 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
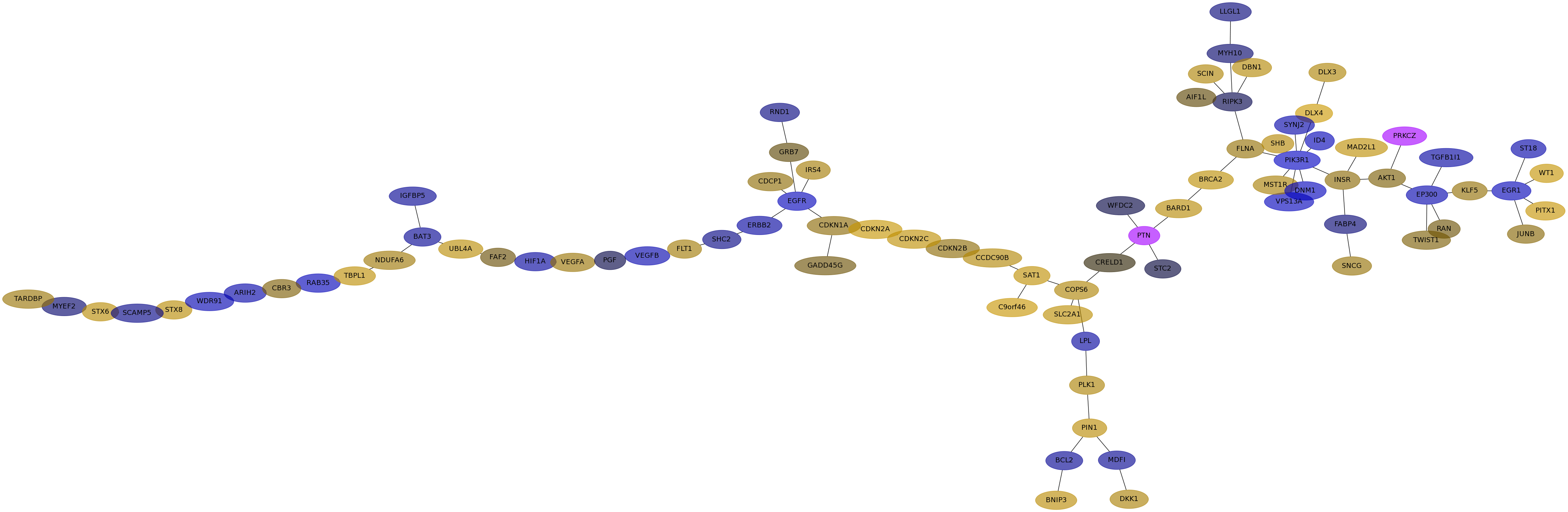
Score for each gene in subnetwork 6937 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| CDCP1 |   | 1 | 1195 | 236 | 292 | 0.068 | 0.045 | -0.058 | undef | 0.010 |
|---|
| FAF2 |   | 1 | 1195 | 236 | 292 | 0.027 | 0.212 | -0.070 | undef | 0.195 |
|---|
| SLC2A1 |   | 7 | 256 | 236 | 237 | 0.205 | 0.143 | -0.050 | -0.023 | 0.086 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| ID4 |   | 35 | 34 | 236 | 223 | -0.179 | 0.137 | 0.241 | undef | 0.036 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| VPS13A |   | 10 | 167 | 236 | 234 | -0.203 | 0.105 | -0.046 | 0.120 | 0.062 |
|---|
| MYH10 |   | 5 | 360 | 236 | 242 | -0.029 | 0.139 | 0.132 | undef | 0.223 |
|---|
| CRELD1 |   | 2 | 743 | 236 | 272 | 0.006 | 0.130 | 0.075 | 0.111 | 0.095 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| MYEF2 |   | 1 | 1195 | 236 | 292 | -0.030 | 0.033 | 0.000 | 0.310 | undef |
|---|
| SHC2 |   | 1 | 1195 | 236 | 292 | -0.053 | 0.284 | -0.036 | 0.022 | 0.078 |
|---|
| SCIN |   | 2 | 743 | 236 | 272 | 0.143 | -0.065 | 0.110 | undef | 0.132 |
|---|
| WFDC2 |   | 5 | 360 | 236 | 242 | -0.011 | -0.011 | 0.333 | -0.162 | -0.157 |
|---|
| CDKN2C |   | 16 | 104 | 236 | 227 | 0.216 | -0.106 | 0.202 | -0.024 | -0.081 |
|---|
| PIN1 |   | 24 | 62 | 179 | 181 | 0.194 | 0.040 | -0.040 | 0.065 | -0.048 |
|---|
| RAB35 |   | 1 | 1195 | 236 | 292 | -0.192 | -0.073 | -0.044 | 0.129 | 0.093 |
|---|
| ST18 |   | 16 | 104 | 236 | 227 | -0.131 | 0.028 | -0.088 | 0.186 | 0.151 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| TARDBP |   | 1 | 1195 | 236 | 292 | 0.090 | 0.196 | 0.068 | -0.019 | -0.018 |
|---|
| RAN |   | 3 | 557 | 236 | 265 | 0.030 | -0.006 | 0.062 | 0.016 | undef |
|---|
| NDUFA6 |   | 1 | 1195 | 236 | 292 | 0.100 | 0.003 | -0.060 | -0.080 | 0.177 |
|---|
| BAT3 |   | 4 | 440 | 236 | 262 | -0.078 | 0.072 | -0.057 | 0.110 | 0.069 |
|---|
| DBN1 |   | 1 | 1195 | 236 | 292 | 0.171 | 0.148 | 0.082 | undef | 0.105 |
|---|
| PLK1 |   | 11 | 148 | 236 | 232 | 0.142 | -0.143 | 0.094 | -0.192 | -0.163 |
|---|
| AKT1 |   | 16 | 104 | 236 | 227 | 0.045 | 0.126 | -0.099 | undef | -0.168 |
|---|
| DLX3 |   | 3 | 557 | 236 | 265 | 0.144 | -0.056 | undef | -0.015 | undef |
|---|
| SAT1 |   | 6 | 301 | 236 | 239 | 0.219 | 0.010 | -0.066 | -0.119 | 0.063 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| PGF |   | 1 | 1195 | 236 | 292 | -0.012 | 0.059 | 0.005 | -0.121 | 0.072 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| MST1R |   | 5 | 360 | 236 | 242 | 0.124 | 0.083 | -0.212 | -0.001 | 0.026 |
|---|
| FLT1 |   | 1 | 1195 | 236 | 292 | 0.109 | 0.069 | 0.124 | 0.124 | 0.177 |
|---|
| MAD2L1 |   | 8 | 222 | 236 | 236 | 0.208 | -0.130 | 0.116 | 0.032 | 0.063 |
|---|
| PRKCZ |   | 2 | 743 | 236 | 272 | undef | 0.180 | -0.141 | -0.038 | -0.036 |
|---|
| C9orf46 |   | 1 | 1195 | 236 | 292 | 0.270 | -0.080 | 0.044 | -0.083 | 0.063 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| LPL |   | 13 | 124 | 236 | 230 | -0.102 | -0.062 | 0.292 | 0.087 | -0.105 |
|---|
| COPS6 |   | 16 | 104 | 1 | 31 | 0.147 | -0.068 | 0.128 | 0.216 | 0.065 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| PTN |   | 24 | 62 | 236 | 225 | undef | 0.022 | 0.000 | 0.054 | -0.092 |
|---|
| FABP4 |   | 6 | 301 | 236 | 239 | -0.036 | -0.028 | 0.227 | 0.072 | 0.107 |
|---|
| SHB |   | 2 | 743 | 236 | 272 | 0.164 | 0.141 | -0.074 | 0.107 | -0.048 |
|---|
| SNCG |   | 3 | 557 | 236 | 265 | 0.080 | -0.086 | 0.088 | -0.090 | -0.226 |
|---|
| UBL4A |   | 1 | 1195 | 236 | 292 | 0.196 | -0.040 | 0.219 | -0.265 | 0.062 |
|---|
| CDKN2B |   | 7 | 256 | 236 | 237 | 0.066 | 0.050 | -0.028 | 0.039 | 0.153 |
|---|
| STX8 |   | 1 | 1195 | 236 | 292 | 0.192 | 0.002 | 0.171 | 0.160 | 0.336 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| CCDC90B |   | 2 | 743 | 236 | 272 | 0.163 | 0.058 | 0.152 | undef | 0.103 |
|---|
| RND1 |   | 6 | 301 | 236 | 239 | -0.046 | 0.085 | 0.022 | 0.082 | 0.113 |
|---|
| STX6 |   | 3 | 557 | 236 | 265 | 0.193 | 0.152 | 0.038 | 0.052 | 0.064 |
|---|
| JUNB |   | 20 | 81 | 179 | 185 | 0.057 | 0.086 | 0.065 | 0.001 | -0.015 |
|---|
| STC2 |   | 5 | 360 | 236 | 242 | -0.009 | 0.056 | 0.181 | -0.185 | 0.016 |
|---|
| LLGL1 |   | 1 | 1195 | 236 | 292 | -0.040 | -0.163 | -0.141 | -0.057 | -0.018 |
|---|
| WT1 |   | 33 | 37 | 1 | 21 | 0.254 | -0.039 | 0.036 | undef | 0.172 |
|---|
| BNIP3 |   | 11 | 148 | 236 | 232 | 0.193 | 0.071 | 0.102 | 0.119 | 0.285 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| SYNJ2 |   | 3 | 557 | 236 | 265 | -0.132 | 0.008 | 0.015 | undef | -0.066 |
|---|
| DLX4 |   | 10 | 167 | 236 | 234 | 0.291 | 0.057 | -0.004 | undef | -0.059 |
|---|
| VEGFB |   | 1 | 1195 | 236 | 292 | -0.146 | -0.014 | 0.079 | 0.196 | -0.105 |
|---|
| MDFI |   | 17 | 95 | 236 | 226 | -0.075 | -0.001 | 0.177 | 0.169 | -0.112 |
|---|
| DNM1 |   | 4 | 440 | 236 | 262 | -0.203 | 0.095 | 0.163 | 0.001 | 0.082 |
|---|
| RIPK3 |   | 19 | 85 | 179 | 186 | -0.014 | 0.040 | undef | 0.173 | undef |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| AIF1L |   | 1 | 1195 | 236 | 292 | 0.021 | -0.099 | undef | undef | undef |
|---|
| IRS4 |   | 32 | 40 | 236 | 224 | 0.115 | -0.111 | -0.088 | 0.096 | 0.142 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| ARIH2 |   | 5 | 360 | 236 | 242 | -0.131 | 0.196 | -0.071 | -0.127 | -0.016 |
|---|
| DKK1 |   | 4 | 440 | 236 | 262 | 0.134 | 0.091 | -0.124 | 0.065 | 0.129 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| HIF1A |   | 13 | 124 | 236 | 230 | -0.098 | 0.224 | -0.118 | 0.135 | 0.356 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| PITX1 |   | 3 | 557 | 236 | 265 | 0.256 | 0.046 | 0.091 | -0.068 | 0.139 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| TBPL1 |   | 3 | 557 | 236 | 265 | 0.211 | -0.037 | 0.023 | 0.061 | 0.217 |
|---|
| SCAMP5 |   | 1 | 1195 | 236 | 292 | -0.056 | 0.019 | undef | undef | undef |
|---|
| CBR3 |   | 1 | 1195 | 236 | 292 | 0.045 | -0.023 | 0.032 | 0.024 | -0.065 |
|---|
| WDR91 |   | 2 | 743 | 236 | 272 | -0.160 | 0.208 | -0.347 | -0.157 | -0.065 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
GO Enrichment output for subnetwork 6937 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.897E-10 | 4.634E-07 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 2.963E-10 | 3.62E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.139E-09 | 9.278E-07 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 1.321E-09 | 8.07E-07 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.822E-09 | 2.356E-06 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.09E-07 | 8.511E-05 |
|---|
| regulation of nitric-oxide synthase activity | GO:0050999 |  | 3.112E-07 | 1.086E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 3.206E-07 | 9.789E-05 |
|---|
| positive regulation of cellular catabolic process | GO:0031331 |  | 4.648E-07 | 1.262E-04 |
|---|
| regulation of monooxygenase activity | GO:0032768 |  | 6.684E-07 | 1.633E-04 |
|---|
| response to UV | GO:0009411 |  | 6.784E-07 | 1.507E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.993E-10 | 7.202E-07 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 3.981E-10 | 4.789E-07 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 1.774E-09 | 1.423E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.792E-09 | 1.078E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.015E-09 | 2.894E-06 |
|---|
| interphase | GO:0051325 |  | 7.933E-09 | 3.181E-06 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.796E-07 | 9.61E-05 |
|---|
| regulation of nitric-oxide synthase activity | GO:0050999 |  | 3.941E-07 | 1.185E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 4.285E-07 | 1.146E-04 |
|---|
| positive regulation of cellular catabolic process | GO:0031331 |  | 5.884E-07 | 1.416E-04 |
|---|
| response to UV | GO:0009411 |  | 7.624E-07 | 1.668E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.185E-09 | 2.726E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.942E-09 | 5.683E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.076E-08 | 1.592E-05 |
|---|
| interphase | GO:0051325 |  | 2.546E-08 | 1.464E-05 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 5.041E-08 | 2.319E-05 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 2.324E-07 | 8.91E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 7.671E-07 | 2.52E-04 |
|---|
| regulation of nitric-oxide synthase activity | GO:0050999 |  | 1.076E-06 | 3.094E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.196E-06 | 3.057E-04 |
|---|
| response to UV | GO:0009411 |  | 1.379E-06 | 3.171E-04 |
|---|
| positive regulation of cellular catabolic process | GO:0031331 |  | 1.605E-06 | 3.355E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell migration | GO:0030335 |  | 4.894E-07 | 9.02E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 8.037E-07 | 7.406E-04 |
|---|
| regulation of cell migration | GO:0030334 |  | 2.054E-06 | 1.262E-03 |
|---|
| regulation of locomotion | GO:0040012 |  | 4.246E-06 | 1.956E-03 |
|---|
| regulation of cell motion | GO:0051270 |  | 4.246E-06 | 1.565E-03 |
|---|
| response to hypoxia | GO:0001666 |  | 5.154E-06 | 1.583E-03 |
|---|
| response to oxygen levels | GO:0070482 |  | 5.76E-06 | 1.517E-03 |
|---|
| positive regulation of glucose metabolic process | GO:0010907 |  | 5.919E-06 | 1.364E-03 |
|---|
| positive regulation of catabolic process | GO:0009896 |  | 8.223E-06 | 1.684E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.041E-05 | 1.918E-03 |
|---|
| regulation of cellular carbohydrate catabolic process | GO:0043471 |  | 1.177E-05 | 1.971E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.185E-09 | 2.726E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.942E-09 | 5.683E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.076E-08 | 1.592E-05 |
|---|
| interphase | GO:0051325 |  | 2.546E-08 | 1.464E-05 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 5.041E-08 | 2.319E-05 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 2.324E-07 | 8.91E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 7.671E-07 | 2.52E-04 |
|---|
| regulation of nitric-oxide synthase activity | GO:0050999 |  | 1.076E-06 | 3.094E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.196E-06 | 3.057E-04 |
|---|
| response to UV | GO:0009411 |  | 1.379E-06 | 3.171E-04 |
|---|
| positive regulation of cellular catabolic process | GO:0031331 |  | 1.605E-06 | 3.355E-04 |
|---|



