Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6878
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7490 | 3.836e-03 | 6.400e-04 | 7.827e-01 | 1.922e-06 |
|---|
| Loi | 0.2309 | 7.928e-02 | 1.110e-02 | 4.258e-01 | 3.748e-04 |
|---|
| Schmidt | 0.6760 | 0.000e+00 | 0.000e+00 | 4.217e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7911 | 0.000e+00 | 0.000e+00 | 3.421e-03 | 0.000e+00 |
|---|
| Wang | 0.2636 | 2.412e-03 | 4.133e-02 | 3.845e-01 | 3.834e-05 |
|---|
Expression data for subnetwork 6878 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
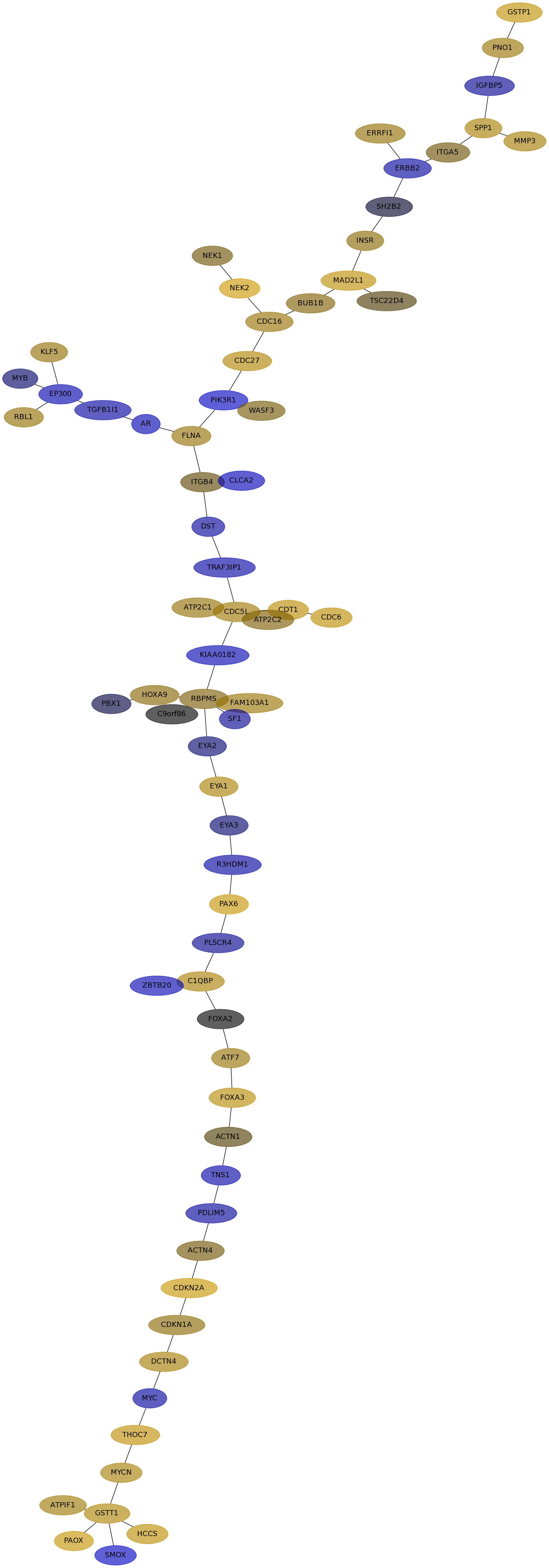
Score for each gene in subnetwork 6878 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| PAX6 |   | 22 | 69 | 318 | 312 | 0.259 | -0.003 | 0.112 | undef | 0.168 |
|---|
| ATP2C2 |   | 1 | 1195 | 1010 | 1045 | 0.047 | 0.102 | -0.097 | 0.118 | -0.029 |
|---|
| PBX1 |   | 6 | 301 | 1 | 52 | -0.010 | 0.145 | -0.013 | 0.176 | -0.053 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| HCCS |   | 1 | 1195 | 1010 | 1045 | 0.205 | -0.068 | -0.021 | 0.120 | 0.053 |
|---|
| DST |   | 22 | 69 | 1 | 25 | -0.094 | 0.343 | 0.065 | -0.062 | 0.194 |
|---|
| RBPMS |   | 3 | 557 | 1010 | 1001 | 0.043 | 0.108 | 0.066 | 0.145 | -0.077 |
|---|
| CDC5L |   | 2 | 743 | 1010 | 1007 | 0.102 | -0.114 | 0.107 | 0.024 | 0.133 |
|---|
| EYA3 |   | 9 | 196 | 318 | 324 | -0.034 | 0.053 | -0.020 | 0.188 | -0.142 |
|---|
| R3HDM1 |   | 4 | 440 | 318 | 337 | -0.108 | -0.192 | -0.049 | 0.082 | 0.223 |
|---|
| CDC6 |   | 3 | 557 | 570 | 572 | 0.198 | -0.053 | 0.038 | -0.202 | 0.095 |
|---|
| ACTN1 |   | 18 | 86 | 318 | 313 | 0.016 | 0.347 | 0.068 | 0.100 | 0.314 |
|---|
| TNS1 |   | 1 | 1195 | 1010 | 1045 | -0.125 | 0.149 | 0.120 | undef | 0.210 |
|---|
| BUB1B |   | 4 | 440 | 1010 | 991 | 0.050 | -0.164 | 0.037 | -0.101 | 0.148 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| PAOX |   | 3 | 557 | 1 | 63 | 0.255 | -0.190 | 0.163 | undef | -0.086 |
|---|
| FOXA2 |   | 2 | 743 | 806 | 806 | 0.000 | -0.024 | -0.078 | undef | 0.153 |
|---|
| CDC16 |   | 2 | 743 | 1010 | 1007 | 0.086 | -0.029 | 0.095 | 0.029 | 0.022 |
|---|
| MMP3 |   | 6 | 301 | 179 | 205 | 0.121 | 0.171 | 0.081 | 0.198 | 0.192 |
|---|
| RBL1 |   | 21 | 73 | 1 | 27 | 0.078 | undef | undef | 0.051 | undef |
|---|
| CLCA2 |   | 9 | 196 | 412 | 411 | -0.176 | 0.243 | -0.153 | 0.191 | -0.000 |
|---|
| HOXA9 |   | 2 | 743 | 1010 | 1007 | 0.057 | 0.136 | 0.040 | undef | 0.018 |
|---|
| MAD2L1 |   | 8 | 222 | 236 | 236 | 0.208 | -0.130 | 0.116 | 0.032 | 0.063 |
|---|
| TRAF3IP1 |   | 9 | 196 | 83 | 99 | -0.113 | -0.063 | -0.088 | 0.276 | undef |
|---|
| ACTN4 |   | 14 | 117 | 318 | 315 | 0.035 | 0.180 | undef | -0.048 | undef |
|---|
| ZBTB20 |   | 1 | 1195 | 1010 | 1045 | -0.163 | 0.269 | -0.045 | 0.056 | 0.014 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| EYA2 |   | 13 | 124 | 318 | 316 | -0.034 | 0.113 | -0.142 | undef | -0.019 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| WASF3 |   | 1 | 1195 | 1010 | 1045 | 0.038 | -0.040 | 0.036 | 0.038 | 0.054 |
|---|
| PLSCR4 |   | 13 | 124 | 318 | 316 | -0.069 | 0.140 | 0.231 | 0.212 | 0.212 |
|---|
| EYA1 |   | 8 | 222 | 1010 | 981 | 0.135 | -0.065 | 0.162 | 0.068 | -0.088 |
|---|
| THOC7 |   | 1 | 1195 | 1010 | 1045 | 0.213 | 0.028 | -0.022 | undef | 0.078 |
|---|
| ATF7 |   | 28 | 47 | 179 | 175 | 0.085 | 0.115 | -0.088 | 0.172 | -0.092 |
|---|
| CDT1 |   | 3 | 557 | 1 | 63 | 0.194 | -0.184 | -0.072 | undef | -0.030 |
|---|
| FOXA3 |   | 12 | 140 | 318 | 322 | 0.192 | 0.289 | undef | -0.171 | undef |
|---|
| GSTT1 |   | 15 | 111 | 1 | 32 | 0.148 | 0.107 | -0.190 | 0.022 | -0.064 |
|---|
| GSTP1 |   | 14 | 117 | 1 | 34 | 0.222 | -0.166 | 0.201 | 0.126 | -0.136 |
|---|
| C1QBP |   | 2 | 743 | 1010 | 1007 | 0.134 | -0.072 | 0.080 | -0.072 | -0.006 |
|---|
| KIAA0182 |   | 1 | 1195 | 1010 | 1045 | -0.163 | 0.155 | -0.377 | 0.174 | 0.050 |
|---|
| PNO1 |   | 3 | 557 | 1010 | 1001 | 0.090 | -0.052 | -0.092 | -0.048 | -0.138 |
|---|
| SPP1 |   | 26 | 52 | 83 | 91 | 0.130 | 0.119 | 0.184 | 0.204 | 0.153 |
|---|
| SMOX |   | 2 | 743 | 1 | 66 | -0.232 | 0.276 | -0.089 | undef | 0.023 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| DCTN4 |   | 5 | 360 | 1010 | 987 | 0.122 | -0.025 | -0.111 | 0.173 | -0.105 |
|---|
| NEK2 |   | 8 | 222 | 141 | 149 | 0.292 | -0.106 | 0.020 | 0.083 | 0.165 |
|---|
| MYCN |   | 7 | 256 | 764 | 756 | 0.125 | 0.057 | -0.003 | -0.081 | -0.084 |
|---|
| ERRFI1 |   | 10 | 167 | 412 | 408 | 0.076 | 0.215 | undef | -0.043 | undef |
|---|
| MYB |   | 28 | 47 | 366 | 344 | -0.028 | 0.075 | 0.002 | 0.065 | -0.012 |
|---|
| ATPIF1 |   | 2 | 743 | 1010 | 1007 | 0.102 | 0.164 | 0.165 | undef | -0.031 |
|---|
| FAM103A1 |   | 1 | 1195 | 1010 | 1045 | 0.087 | -0.129 | undef | undef | undef |
|---|
| SF1 |   | 1 | 1195 | 1010 | 1045 | -0.079 | 0.119 | 0.188 | 0.164 | -0.032 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| ATP2C1 |   | 2 | 743 | 1010 | 1007 | 0.081 | 0.198 | 0.081 | -0.041 | 0.120 |
|---|
| ITGA5 |   | 26 | 52 | 179 | 177 | 0.031 | 0.192 | 0.125 | 0.133 | 0.202 |
|---|
| NEK1 |   | 15 | 111 | 141 | 139 | 0.032 | 0.184 | 0.049 | undef | 0.135 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| CDC27 |   | 6 | 301 | 1010 | 986 | 0.156 | -0.086 | -0.029 | 0.006 | 0.348 |
|---|
| SH2B2 |   | 2 | 743 | 1010 | 1007 | -0.006 | -0.062 | -0.069 | -0.056 | -0.111 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| C9orf86 |   | 2 | 743 | 1010 | 1007 | -0.000 | 0.023 | -0.199 | undef | 0.097 |
|---|
| TSC22D4 |   | 1 | 1195 | 1010 | 1045 | 0.015 | -0.213 | -0.002 | undef | -0.055 |
|---|
| ITGB4 |   | 35 | 34 | 1 | 20 | 0.021 | 0.313 | -0.134 | 0.080 | -0.114 |
|---|
| PDLIM5 |   | 4 | 440 | 318 | 337 | -0.091 | 0.242 | -0.085 | -0.001 | 0.117 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
GO Enrichment output for subnetwork 6878 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| sex determination | GO:0007530 |  | 1.62E-07 | 3.959E-04 |
|---|
| C21-steroid hormone biosynthetic process | GO:0006700 |  | 1.369E-06 | 1.672E-03 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 1.791E-06 | 1.459E-03 |
|---|
| cell-substrate junction assembly | GO:0007044 |  | 2.283E-06 | 1.394E-03 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 2.313E-06 | 1.13E-03 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 4.333E-06 | 1.764E-03 |
|---|
| C21-steroid hormone metabolic process | GO:0008207 |  | 4.409E-06 | 1.539E-03 |
|---|
| regulation of ligase activity | GO:0051340 |  | 6.198E-06 | 1.893E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 8.255E-06 | 2.241E-03 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 8.255E-06 | 2.017E-03 |
|---|
| negative regulation of DNA replication initiation | GO:0032297 |  | 8.255E-06 | 1.833E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| sex determination | GO:0007530 |  | 2.163E-07 | 5.205E-04 |
|---|
| C21-steroid hormone biosynthetic process | GO:0006700 |  | 1.565E-06 | 1.883E-03 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 3.268E-06 | 2.621E-03 |
|---|
| cell-substrate junction assembly | GO:0007044 |  | 3.46E-06 | 2.081E-03 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 3.562E-06 | 1.714E-03 |
|---|
| C21-steroid hormone metabolic process | GO:0008207 |  | 5.427E-06 | 2.176E-03 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 7.863E-06 | 2.703E-03 |
|---|
| regulation of ligase activity | GO:0051340 |  | 1.122E-05 | 3.374E-03 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 1.131E-05 | 3.024E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.131E-05 | 2.721E-03 |
|---|
| negative regulation of DNA replication initiation | GO:0032297 |  | 1.131E-05 | 2.474E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| sex determination | GO:0007530 |  | 2.523E-07 | 5.802E-04 |
|---|
| C21-steroid hormone biosynthetic process | GO:0006700 |  | 3.021E-06 | 3.474E-03 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 6.821E-06 | 5.229E-03 |
|---|
| cell-substrate junction assembly | GO:0007044 |  | 7.057E-06 | 4.058E-03 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 9.04E-06 | 4.158E-03 |
|---|
| C21-steroid hormone metabolic process | GO:0008207 |  | 1.136E-05 | 4.354E-03 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 1.653E-05 | 5.432E-03 |
|---|
| regulation of ligase activity | GO:0051340 |  | 2.267E-05 | 6.519E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.344E-05 | 5.991E-03 |
|---|
| middle ear morphogenesis | GO:0042474 |  | 2.344E-05 | 5.392E-03 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 2.344E-05 | 4.902E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| actin cytoskeleton reorganization | GO:0031532 |  | 2.772E-06 | 5.108E-03 |
|---|
| sex determination | GO:0007530 |  | 1.53E-05 | 0.01410351 |
|---|
| regulation of mitotic cell cycle | GO:0007346 |  | 8.733E-05 | 0.05364939 |
|---|
| regulation of mitosis | GO:0007088 |  | 8.812E-05 | 0.04060212 |
|---|
| integrin-mediated signaling pathway | GO:0007229 |  | 8.812E-05 | 0.03248169 |
|---|
| wound healing | GO:0042060 |  | 9.358E-05 | 0.02874479 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 1.626E-04 | 0.0428134 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.626E-04 | 0.03746173 |
|---|
| regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051439 |  | 2.325E-04 | 0.04760906 |
|---|
| multicellular organismal metabolic process | GO:0044236 |  | 2.515E-04 | 0.04635349 |
|---|
| heart development | GO:0007507 |  | 2.625E-04 | 0.04398693 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| sex determination | GO:0007530 |  | 2.523E-07 | 5.802E-04 |
|---|
| C21-steroid hormone biosynthetic process | GO:0006700 |  | 3.021E-06 | 3.474E-03 |
|---|
| negative regulation of ligase activity | GO:0051352 |  | 6.821E-06 | 5.229E-03 |
|---|
| cell-substrate junction assembly | GO:0007044 |  | 7.057E-06 | 4.058E-03 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 9.04E-06 | 4.158E-03 |
|---|
| C21-steroid hormone metabolic process | GO:0008207 |  | 1.136E-05 | 4.354E-03 |
|---|
| regulation of ubiquitin-protein ligase activity | GO:0051438 |  | 1.653E-05 | 5.432E-03 |
|---|
| regulation of ligase activity | GO:0051340 |  | 2.267E-05 | 6.519E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.344E-05 | 5.991E-03 |
|---|
| middle ear morphogenesis | GO:0042474 |  | 2.344E-05 | 5.392E-03 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 2.344E-05 | 4.902E-03 |
|---|



