Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6830
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7496 | 3.795e-03 | 6.310e-04 | 7.813e-01 | 1.871e-06 |
|---|
| Loi | 0.2308 | 7.949e-02 | 1.115e-02 | 4.263e-01 | 3.780e-04 |
|---|
| Schmidt | 0.6757 | 0.000e+00 | 0.000e+00 | 4.252e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7910 | 0.000e+00 | 0.000e+00 | 3.434e-03 | 0.000e+00 |
|---|
| Wang | 0.2636 | 2.423e-03 | 4.143e-02 | 3.850e-01 | 3.866e-05 |
|---|
Expression data for subnetwork 6830 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
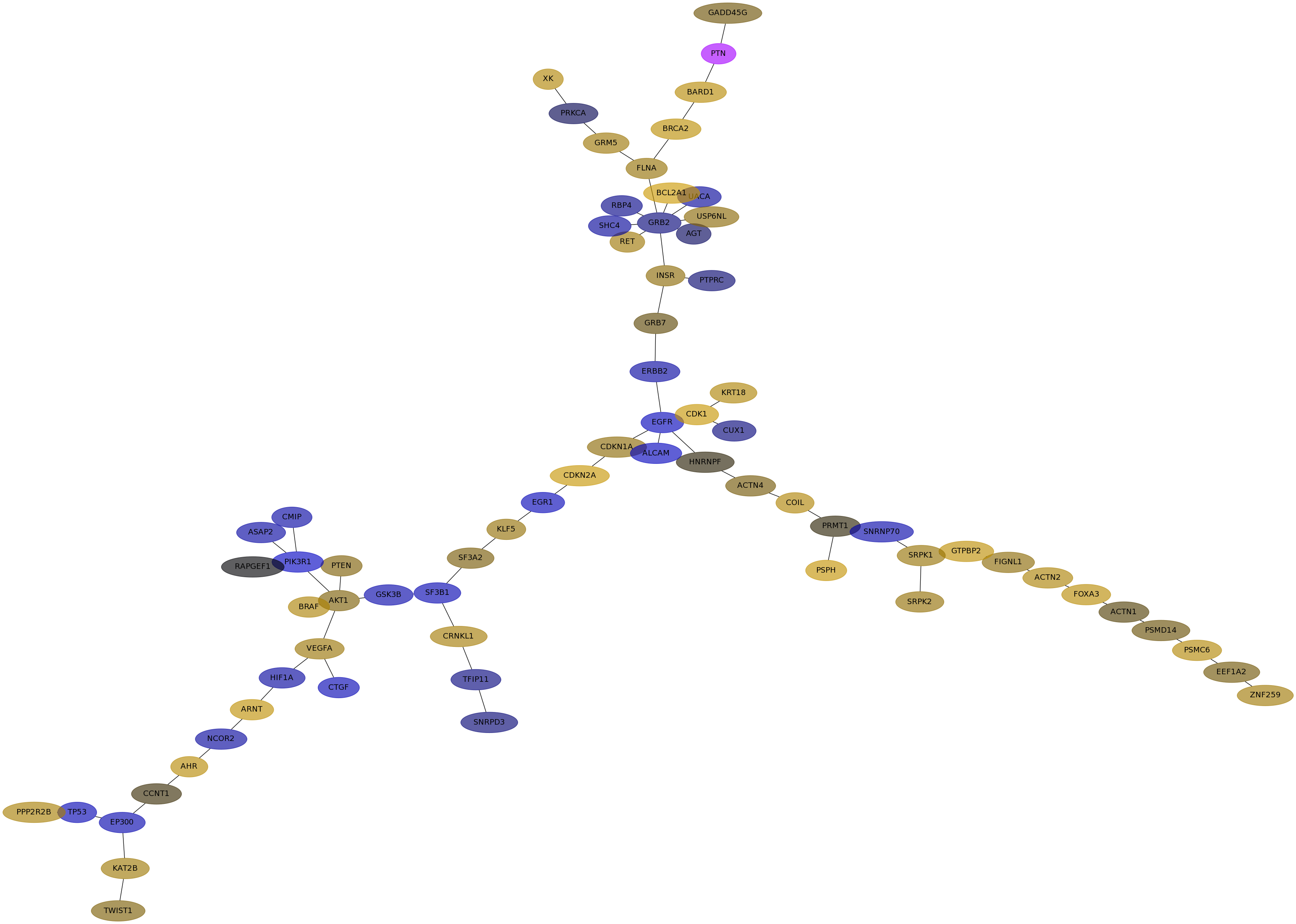
Score for each gene in subnetwork 6830 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| TFIP11 |   | 1 | 1195 | 1203 | 1226 | -0.045 | 0.111 | -0.014 | undef | 0.039 |
|---|
| SF3B1 |   | 18 | 86 | 296 | 280 | -0.154 | 0.126 | -0.073 | -0.131 | -0.204 |
|---|
| RET |   | 7 | 256 | 366 | 376 | 0.098 | 0.063 | -0.179 | undef | -0.159 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| USP6NL |   | 2 | 743 | 1203 | 1206 | 0.060 | -0.111 | undef | undef | undef |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| SNRPD3 |   | 1 | 1195 | 1203 | 1226 | -0.036 | -0.063 | 0.099 | -0.077 | -0.143 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| GRM5 |   | 15 | 111 | 179 | 191 | 0.094 | -0.019 | 0.000 | 0.093 | -0.093 |
|---|
| ASAP2 |   | 1 | 1195 | 1203 | 1226 | -0.110 | 0.001 | 0.103 | -0.007 | -0.027 |
|---|
| GTPBP2 |   | 2 | 743 | 1203 | 1206 | 0.211 | 0.033 | -0.132 | 0.119 | -0.002 |
|---|
| CTGF |   | 32 | 40 | 318 | 290 | -0.169 | 0.181 | 0.126 | 0.101 | 0.209 |
|---|
| ZNF259 |   | 1 | 1195 | 1203 | 1226 | 0.103 | 0.099 | -0.062 | -0.022 | 0.112 |
|---|
| PSMD14 |   | 4 | 440 | 842 | 831 | 0.027 | 0.135 | -0.179 | -0.018 | -0.031 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| SRPK2 |   | 3 | 557 | 1203 | 1187 | 0.076 | 0.183 | -0.087 | undef | 0.054 |
|---|
| AKT1 |   | 16 | 104 | 236 | 227 | 0.045 | 0.126 | -0.099 | undef | -0.168 |
|---|
| CMIP |   | 1 | 1195 | 1203 | 1226 | -0.112 | 0.009 | undef | undef | undef |
|---|
| FIGNL1 |   | 1 | 1195 | 1203 | 1226 | 0.064 | -0.193 | undef | undef | undef |
|---|
| CUX1 |   | 6 | 301 | 1203 | 1169 | -0.042 | 0.256 | -0.121 | 0.105 | 0.040 |
|---|
| HNRNPF |   | 2 | 743 | 806 | 806 | 0.005 | -0.112 | 0.124 | 0.001 | -0.055 |
|---|
| CRNKL1 |   | 2 | 743 | 478 | 490 | 0.115 | -0.056 | -0.222 | 0.124 | 0.084 |
|---|
| ACTN1 |   | 18 | 86 | 318 | 313 | 0.016 | 0.347 | 0.068 | 0.100 | 0.314 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| ALCAM |   | 8 | 222 | 1 | 46 | -0.208 | 0.193 | 0.067 | 0.088 | -0.010 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| RBP4 |   | 4 | 440 | 764 | 761 | -0.059 | -0.011 | 0.206 | 0.334 | -0.045 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| CCNT1 |   | 6 | 301 | 478 | 473 | 0.008 | -0.028 | 0.162 | 0.007 | -0.213 |
|---|
| PSMC6 |   | 5 | 360 | 318 | 334 | 0.163 | 0.061 | -0.010 | 0.069 | 0.044 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| ACTN4 |   | 14 | 117 | 318 | 315 | 0.035 | 0.180 | undef | -0.048 | undef |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| COIL |   | 4 | 440 | 913 | 904 | 0.151 | 0.144 | 0.071 | 0.106 | 0.044 |
|---|
| ARNT |   | 4 | 440 | 1203 | 1184 | 0.221 | -0.023 | 0.140 | 0.122 | -0.124 |
|---|
| PTN |   | 24 | 62 | 236 | 225 | undef | 0.022 | 0.000 | 0.054 | -0.092 |
|---|
| SRPK1 |   | 2 | 743 | 1203 | 1206 | 0.085 | -0.010 | 0.143 | 0.037 | 0.191 |
|---|
| PSPH |   | 20 | 81 | 366 | 356 | 0.240 | -0.019 | -0.102 | 0.358 | 0.025 |
|---|
| NCOR2 |   | 32 | 40 | 318 | 290 | -0.096 | 0.211 | -0.163 | 0.158 | 0.114 |
|---|
| SHC4 |   | 1 | 1195 | 1203 | 1226 | -0.088 | 0.034 | undef | undef | undef |
|---|
| BRAF |   | 1 | 1195 | 1203 | 1226 | 0.139 | -0.115 | -0.115 | -0.049 | -0.083 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| KAT2B |   | 13 | 124 | 318 | 316 | 0.102 | -0.157 | -0.091 | 0.136 | -0.087 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| PRMT1 |   | 2 | 743 | 1203 | 1206 | 0.006 | -0.136 | 0.075 | -0.144 | -0.165 |
|---|
| FOXA3 |   | 12 | 140 | 318 | 322 | 0.192 | 0.289 | undef | -0.171 | undef |
|---|
| ACTN2 |   | 2 | 743 | 806 | 806 | 0.139 | -0.106 | 0.155 | undef | 0.127 |
|---|
| PTEN |   | 27 | 50 | 141 | 135 | 0.045 | 0.055 | 0.017 | 0.183 | -0.041 |
|---|
| EEF1A2 |   | 8 | 222 | 457 | 450 | 0.032 | 0.220 | -0.119 | 0.018 | 0.102 |
|---|
| RAPGEF1 |   | 1 | 1195 | 1203 | 1226 | -0.000 | -0.093 | 0.126 | -0.008 | -0.119 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| SF3A2 |   | 3 | 557 | 1203 | 1187 | 0.038 | -0.181 | 0.094 | -0.172 | -0.067 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| XK |   | 18 | 86 | 179 | 187 | 0.154 | -0.000 | 0.067 | -0.123 | -0.040 |
|---|
| PTPRC |   | 9 | 196 | 83 | 99 | -0.031 | -0.204 | -0.050 | -0.179 | -0.068 |
|---|
| GRB2 |   | 13 | 124 | 318 | 316 | -0.040 | -0.159 | -0.036 | -0.028 | -0.024 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| HIF1A |   | 13 | 124 | 236 | 230 | -0.098 | 0.224 | -0.118 | 0.135 | 0.356 |
|---|
| BCL2A1 |   | 4 | 440 | 764 | 761 | 0.298 | -0.120 | -0.087 | -0.199 | -0.043 |
|---|
| AGT |   | 4 | 440 | 1203 | 1184 | -0.020 | -0.101 | -0.043 | 0.255 | 0.064 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| UACA |   | 1 | 1195 | 1203 | 1226 | -0.088 | 0.089 | undef | undef | undef |
|---|
| SNRNP70 |   | 5 | 360 | 296 | 310 | -0.129 | 0.130 | -0.059 | -0.099 | 0.036 |
|---|
| KRT18 |   | 8 | 222 | 590 | 569 | 0.148 | 0.100 | -0.083 | 0.024 | -0.053 |
|---|
| AHR |   | 8 | 222 | 478 | 461 | 0.180 | 0.067 | 0.091 | -0.182 | -0.129 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
GO Enrichment output for subnetwork 6830 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.307E-11 | 3.194E-08 |
|---|
| response to UV | GO:0009411 |  | 3.676E-10 | 4.491E-07 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 1.42E-09 | 1.156E-06 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.936E-09 | 1.182E-06 |
|---|
| lung development | GO:0030324 |  | 2.377E-09 | 1.161E-06 |
|---|
| positive regulation of cellular catabolic process | GO:0031331 |  | 2.424E-09 | 9.87E-07 |
|---|
| glutathione biosynthetic process | GO:0006750 |  | 2.424E-09 | 8.46E-07 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.931E-09 | 8.95E-07 |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 3.168E-09 | 8.599E-07 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 3.168E-09 | 7.739E-07 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 3.283E-09 | 7.292E-07 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.563E-11 | 6.168E-08 |
|---|
| response to UV | GO:0009411 |  | 4.918E-10 | 5.916E-07 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 2.089E-09 | 1.676E-06 |
|---|
| glutathione biosynthetic process | GO:0006750 |  | 2.089E-09 | 1.257E-06 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 3.061E-09 | 1.473E-06 |
|---|
| lung development | GO:0030324 |  | 3.392E-09 | 1.36E-06 |
|---|
| positive regulation of cellular catabolic process | GO:0031331 |  | 3.566E-09 | 1.226E-06 |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 4.325E-09 | 1.301E-06 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 4.325E-09 | 1.156E-06 |
|---|
| respiratory tube development | GO:0030323 |  | 4.841E-09 | 1.165E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.966E-09 | 1.086E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.812E-09 | 4.168E-06 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 8.046E-09 | 9.253E-06 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 1.015E-08 | 7.783E-06 |
|---|
| positive regulation of cellular catabolic process | GO:0031331 |  | 1.024E-08 | 5.887E-06 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.396E-08 | 6.422E-06 |
|---|
| positive regulation of catabolic process | GO:0009896 |  | 2.921E-08 | 1.12E-05 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 3.029E-08 | 9.954E-06 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.131E-07 | 3.251E-05 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.279E-07 | 3.268E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.399E-07 | 3.217E-05 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 1.399E-07 | 2.925E-05 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.031E-08 | 1.9E-05 |
|---|
| regulation of cell motion | GO:0051270 |  | 9.009E-08 | 8.302E-05 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 2.016E-07 | 1.238E-04 |
|---|
| glucose homeostasis | GO:0042593 |  | 7E-07 | 3.225E-04 |
|---|
| regulation of cell migration | GO:0030334 |  | 7.443E-07 | 2.744E-04 |
|---|
| regulation of locomotion | GO:0040012 |  | 1.551E-06 | 4.764E-04 |
|---|
| heart development | GO:0007507 |  | 2.989E-06 | 7.868E-04 |
|---|
| positive regulation of glucose metabolic process | GO:0010907 |  | 3.798E-06 | 8.751E-04 |
|---|
| positive regulation of catabolic process | GO:0009896 |  | 4.566E-06 | 9.35E-04 |
|---|
| tissue remodeling | GO:0048771 |  | 5.938E-06 | 1.094E-03 |
|---|
| regulation of oxidoreductase activity | GO:0051341 |  | 5.938E-06 | 9.949E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.812E-09 | 4.168E-06 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 8.046E-09 | 9.253E-06 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 1.015E-08 | 7.783E-06 |
|---|
| positive regulation of cellular catabolic process | GO:0031331 |  | 1.024E-08 | 5.887E-06 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.396E-08 | 6.422E-06 |
|---|
| positive regulation of catabolic process | GO:0009896 |  | 2.921E-08 | 1.12E-05 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 3.029E-08 | 9.954E-06 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.131E-07 | 3.251E-05 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.279E-07 | 3.268E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.399E-07 | 3.217E-05 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 1.399E-07 | 2.925E-05 |
|---|



