Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6827
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7496 | 3.794e-03 | 6.310e-04 | 7.812e-01 | 1.870e-06 |
|---|
| Loi | 0.2308 | 7.957e-02 | 1.118e-02 | 4.265e-01 | 3.793e-04 |
|---|
| Schmidt | 0.6757 | 0.000e+00 | 0.000e+00 | 4.249e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7910 | 0.000e+00 | 0.000e+00 | 3.433e-03 | 0.000e+00 |
|---|
| Wang | 0.2636 | 2.417e-03 | 4.138e-02 | 3.848e-01 | 3.849e-05 |
|---|
Expression data for subnetwork 6827 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
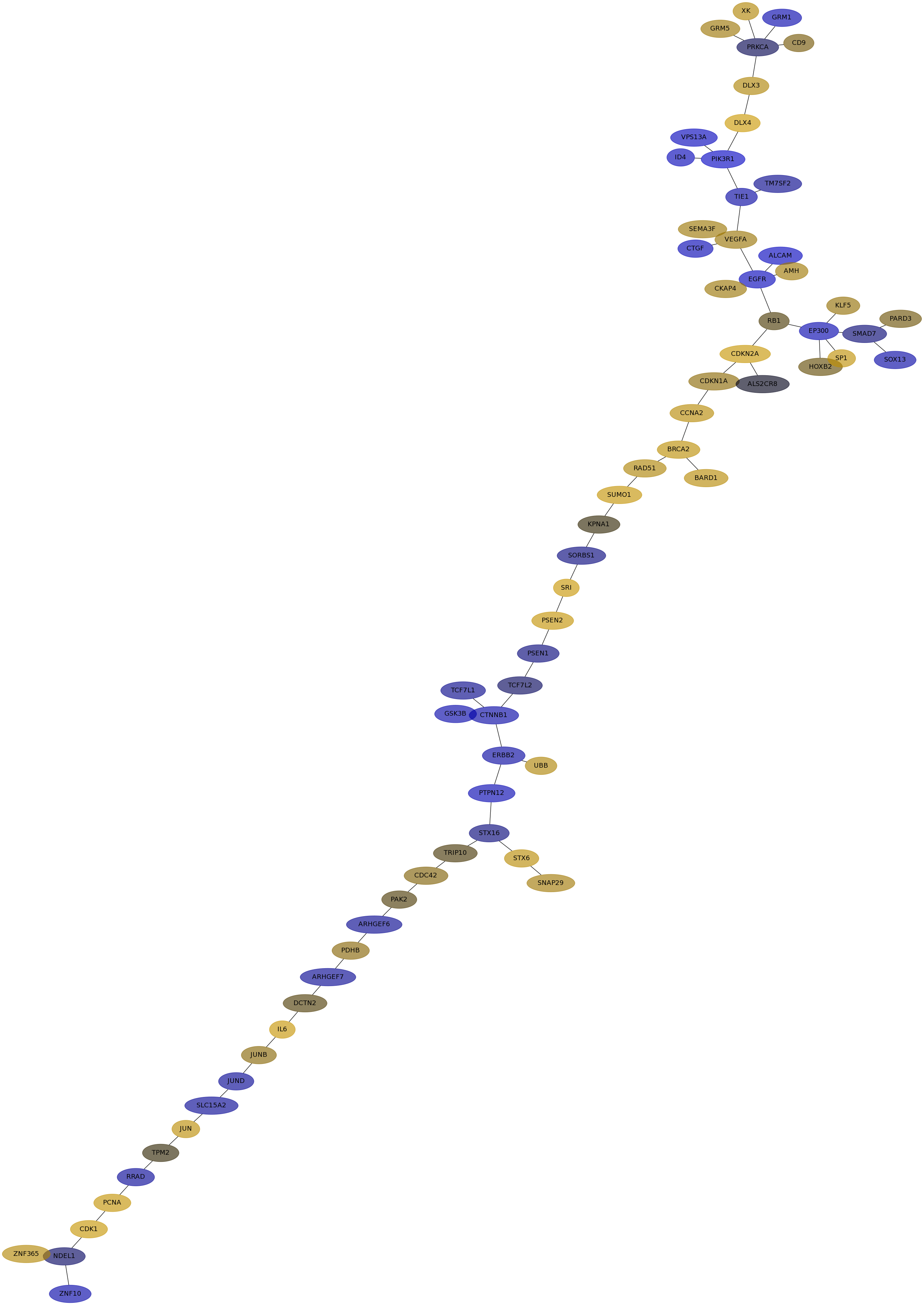
Score for each gene in subnetwork 6827 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| TRIP10 |   | 5 | 360 | 1034 | 1004 | 0.012 | 0.154 | 0.049 | undef | 0.144 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| ID4 |   | 35 | 34 | 236 | 223 | -0.179 | 0.137 | 0.241 | undef | 0.036 |
|---|
| RRAD |   | 2 | 743 | 1034 | 1039 | -0.081 | 0.171 | -0.082 | 0.069 | 0.117 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| VPS13A |   | 10 | 167 | 236 | 234 | -0.203 | 0.105 | -0.046 | 0.120 | 0.062 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| GRM5 |   | 15 | 111 | 179 | 191 | 0.094 | -0.019 | 0.000 | 0.093 | -0.093 |
|---|
| JUND |   | 2 | 743 | 991 | 999 | -0.077 | 0.091 | -0.075 | 0.007 | -0.005 |
|---|
| PCNA |   | 15 | 111 | 1 | 32 | 0.243 | -0.158 | 0.027 | -0.099 | 0.003 |
|---|
| CCNA2 |   | 21 | 73 | 179 | 183 | 0.176 | -0.053 | 0.188 | -0.078 | 0.246 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| CTGF |   | 32 | 40 | 318 | 290 | -0.169 | 0.181 | 0.126 | 0.101 | 0.209 |
|---|
| CDC42 |   | 26 | 52 | 179 | 177 | 0.047 | 0.242 | -0.027 | 0.040 | -0.042 |
|---|
| SRI |   | 7 | 256 | 614 | 601 | 0.270 | -0.040 | 0.181 | 0.185 | 0.057 |
|---|
| PSEN2 |   | 14 | 117 | 614 | 591 | 0.239 | -0.068 | -0.157 | 0.295 | 0.045 |
|---|
| ZNF365 |   | 4 | 440 | 141 | 165 | 0.161 | 0.190 | -0.050 | -0.053 | -0.008 |
|---|
| NDEL1 |   | 1 | 1195 | 1034 | 1069 | -0.023 | 0.211 | 0.146 | undef | 0.221 |
|---|
| DLX3 |   | 3 | 557 | 236 | 265 | 0.144 | -0.056 | undef | -0.015 | undef |
|---|
| SEMA3F |   | 2 | 743 | 83 | 123 | 0.097 | 0.035 | 0.094 | undef | -0.068 |
|---|
| ZNF10 |   | 1 | 1195 | 1034 | 1069 | -0.125 | 0.125 | -0.014 | undef | 0.014 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| ALCAM |   | 8 | 222 | 1 | 46 | -0.208 | 0.193 | 0.067 | 0.088 | -0.010 |
|---|
| STX16 |   | 2 | 743 | 1034 | 1039 | -0.041 | 0.027 | 0.279 | 0.073 | 0.088 |
|---|
| PDHB |   | 13 | 124 | 525 | 503 | 0.056 | 0.017 | 0.197 | -0.205 | 0.193 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| TCF7L1 |   | 2 | 743 | 1034 | 1039 | -0.062 | -0.031 | 0.211 | undef | -0.108 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| ARHGEF6 |   | 10 | 167 | 525 | 507 | -0.068 | -0.120 | 0.166 | undef | -0.115 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| PARD3 |   | 6 | 301 | 366 | 377 | 0.030 | -0.006 | -0.052 | 0.040 | -0.090 |
|---|
| ARHGEF7 |   | 25 | 59 | 83 | 92 | -0.074 | -0.061 | 0.208 | 0.028 | 0.015 |
|---|
| DCTN2 |   | 8 | 222 | 366 | 373 | 0.014 | 0.150 | 0.061 | 0.113 | 0.303 |
|---|
| SMAD7 |   | 5 | 360 | 1034 | 1004 | -0.035 | 0.131 | 0.169 | 0.061 | 0.147 |
|---|
| PSEN1 |   | 10 | 167 | 366 | 362 | -0.041 | 0.110 | undef | 0.083 | undef |
|---|
| HOXB2 |   | 33 | 37 | 318 | 289 | 0.024 | -0.143 | 0.076 | 0.247 | -0.184 |
|---|
| SUMO1 |   | 5 | 360 | 1034 | 1004 | 0.245 | -0.007 | 0.144 | 0.083 | 0.118 |
|---|
| STX6 |   | 3 | 557 | 236 | 265 | 0.193 | 0.152 | 0.038 | 0.052 | 0.064 |
|---|
| JUN |   | 26 | 52 | 179 | 177 | 0.200 | 0.051 | 0.220 | undef | 0.200 |
|---|
| GRM1 |   | 2 | 743 | 1034 | 1039 | -0.140 | -0.030 | 0.000 | 0.093 | -0.105 |
|---|
| RB1 |   | 31 | 43 | 1 | 22 | 0.013 | -0.051 | 0.057 | 0.100 | -0.082 |
|---|
| AMH |   | 10 | 167 | 366 | 362 | 0.103 | 0.056 | 0.113 | -0.187 | 0.301 |
|---|
| PAK2 |   | 26 | 52 | 179 | 177 | 0.014 | 0.161 | 0.081 | -0.044 | 0.062 |
|---|
| JUNB |   | 20 | 81 | 179 | 185 | 0.057 | 0.086 | 0.065 | 0.001 | -0.015 |
|---|
| CD9 |   | 13 | 124 | 525 | 503 | 0.036 | -0.115 | -0.109 | 0.281 | -0.147 |
|---|
| ALS2CR8 |   | 1 | 1195 | 1034 | 1069 | -0.003 | 0.065 | 0.140 | undef | -0.092 |
|---|
| UBB |   | 3 | 557 | 696 | 695 | 0.146 | -0.027 | 0.012 | undef | -0.019 |
|---|
| DLX4 |   | 10 | 167 | 236 | 234 | 0.291 | 0.057 | -0.004 | undef | -0.059 |
|---|
| CKAP4 |   | 6 | 301 | 780 | 764 | 0.093 | 0.152 | -0.167 | 0.315 | -0.017 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| TPM2 |   | 4 | 440 | 1034 | 1015 | 0.007 | 0.269 | 0.212 | 0.051 | 0.203 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| TM7SF2 |   | 2 | 743 | 590 | 616 | -0.064 | -0.016 | 0.022 | 0.106 | -0.048 |
|---|
| PTPN12 |   | 6 | 301 | 478 | 473 | -0.159 | 0.153 | 0.031 | 0.083 | 0.042 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| SLC15A2 |   | 4 | 440 | 928 | 912 | -0.076 | 0.109 | -0.068 | 0.050 | -0.123 |
|---|
| TCF7L2 |   | 17 | 95 | 366 | 358 | -0.021 | 0.143 | 0.280 | undef | 0.101 |
|---|
| XK |   | 18 | 86 | 179 | 187 | 0.154 | -0.000 | 0.067 | -0.123 | -0.040 |
|---|
| SOX13 |   | 4 | 440 | 1034 | 1015 | -0.113 | 0.095 | -0.002 | 0.271 | -0.184 |
|---|
| KPNA1 |   | 9 | 196 | 141 | 147 | 0.007 | 0.152 | -0.083 | 0.073 | 0.086 |
|---|
| RAD51 |   | 13 | 124 | 727 | 699 | 0.146 | -0.098 | 0.135 | -0.070 | 0.061 |
|---|
| SNAP29 |   | 1 | 1195 | 1034 | 1069 | 0.105 | 0.161 | 0.064 | -0.032 | 0.062 |
|---|
| IL6 |   | 9 | 196 | 366 | 365 | 0.257 | -0.106 | 0.226 | 0.024 | 0.035 |
|---|
| TIE1 |   | 18 | 86 | 366 | 357 | -0.108 | 0.005 | 0.141 | -0.108 | 0.252 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
GO Enrichment output for subnetwork 6827 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.071E-11 | 2.616E-08 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.941E-09 | 3.593E-06 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 4.986E-09 | 4.06E-06 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 5.799E-08 | 3.542E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.013E-07 | 4.949E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.039E-07 | 4.232E-05 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 1.095E-07 | 3.821E-05 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.12E-07 | 3.421E-05 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 1.393E-07 | 3.78E-05 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 1.651E-07 | 4.034E-05 |
|---|
| protein sumoylation | GO:0016925 |  | 1.725E-07 | 3.831E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 6.55E-12 | 1.576E-08 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 3.929E-09 | 4.726E-06 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 6.657E-09 | 5.339E-06 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 3.548E-08 | 2.134E-05 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 1.083E-07 | 5.211E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.143E-07 | 4.582E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.266E-07 | 4.351E-05 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.268E-07 | 3.812E-05 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 1.395E-07 | 3.73E-05 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 1.89E-07 | 4.546E-05 |
|---|
| protein sumoylation | GO:0016925 |  | 2.1E-07 | 4.594E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.571E-08 | 3.614E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 2.722E-08 | 3.131E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.101E-07 | 8.441E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 2.189E-07 | 1.259E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 2.189E-07 | 1.007E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 2.424E-07 | 9.294E-05 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.74E-07 | 9.002E-05 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 3.343E-07 | 9.611E-05 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 3.901E-07 | 9.969E-05 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 4.361E-07 | 1.003E-04 |
|---|
| regulation of transcription factor activity | GO:0051090 |  | 4.885E-07 | 1.021E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| membrane fusion | GO:0006944 |  | 1.595E-06 | 2.94E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.668E-06 | 1.537E-03 |
|---|
| amyloid precursor protein catabolic process | GO:0042987 |  | 2.772E-06 | 1.703E-03 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 5.518E-06 | 2.542E-03 |
|---|
| amyloid precursor protein metabolic process | GO:0042982 |  | 9.611E-06 | 3.542E-03 |
|---|
| membrane protein ectodomain proteolysis | GO:0006509 |  | 9.611E-06 | 2.952E-03 |
|---|
| platelet activation | GO:0030168 |  | 9.611E-06 | 2.53E-03 |
|---|
| plasma membrane fusion | GO:0045026 |  | 1.53E-05 | 3.526E-03 |
|---|
| positive regulation of leukocyte migration | GO:0002687 |  | 1.53E-05 | 3.134E-03 |
|---|
| regulation of leukocyte migration | GO:0002685 |  | 2.285E-05 | 4.211E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 3.185E-05 | 5.337E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.571E-08 | 3.614E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 2.722E-08 | 3.131E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.101E-07 | 8.441E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 2.189E-07 | 1.259E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 2.189E-07 | 1.007E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 2.424E-07 | 9.294E-05 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.74E-07 | 9.002E-05 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 3.343E-07 | 9.611E-05 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 3.901E-07 | 9.969E-05 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 4.361E-07 | 1.003E-04 |
|---|
| regulation of transcription factor activity | GO:0051090 |  | 4.885E-07 | 1.021E-04 |
|---|



