Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6808
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7498 | 3.778e-03 | 6.270e-04 | 7.806e-01 | 1.849e-06 |
|---|
| Loi | 0.2308 | 7.947e-02 | 1.115e-02 | 4.263e-01 | 3.778e-04 |
|---|
| Schmidt | 0.6755 | 0.000e+00 | 0.000e+00 | 4.272e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7910 | 0.000e+00 | 0.000e+00 | 3.435e-03 | 0.000e+00 |
|---|
| Wang | 0.2634 | 2.439e-03 | 4.159e-02 | 3.858e-01 | 3.913e-05 |
|---|
Expression data for subnetwork 6808 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
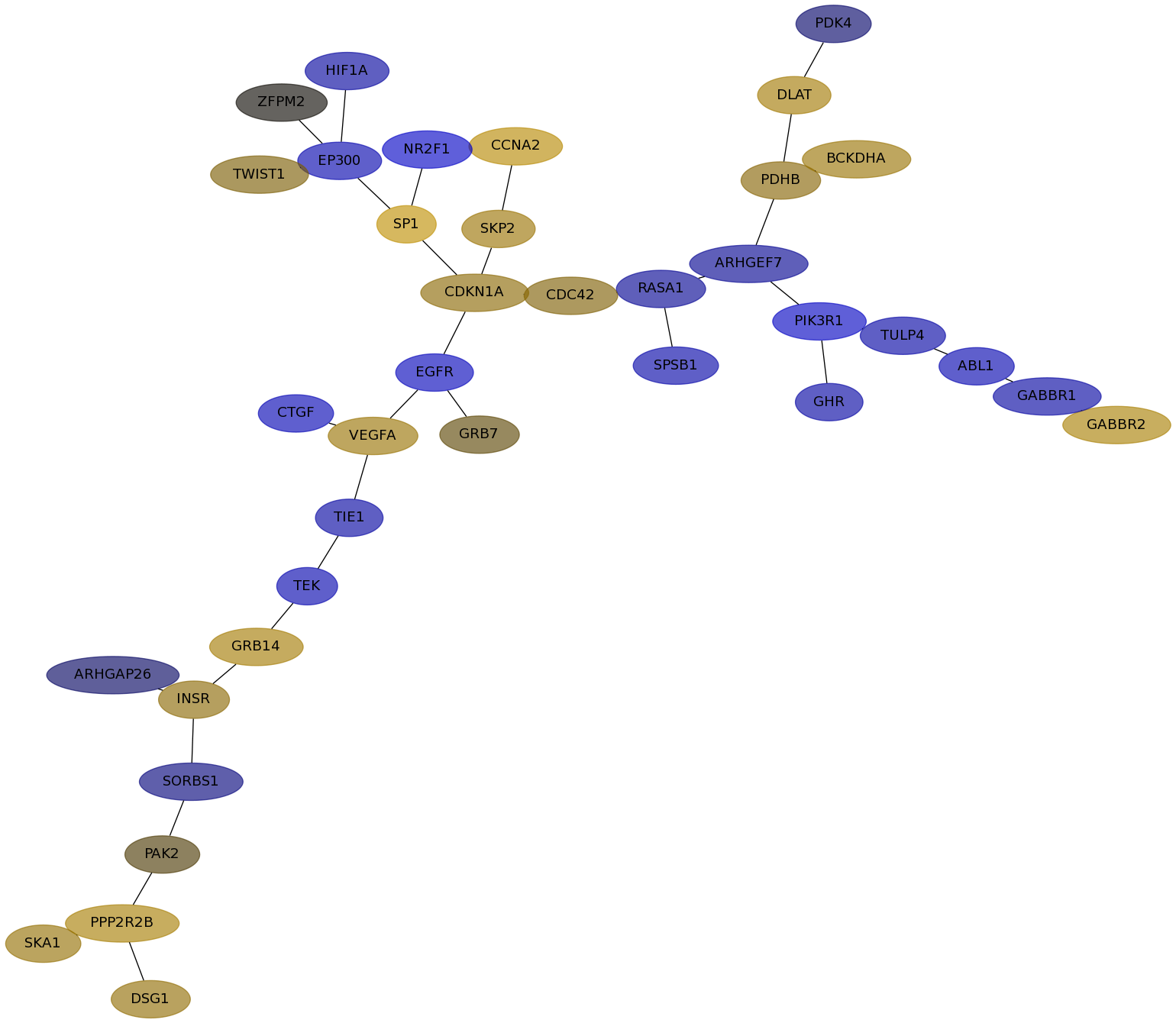
Score for each gene in subnetwork 6808 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| TEK |   | 18 | 86 | 83 | 95 | -0.149 | -0.049 | 0.174 | 0.015 | 0.113 |
|---|
| ARHGAP26 |   | 21 | 73 | 457 | 438 | -0.023 | 0.171 | -0.222 | 0.015 | -0.348 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| GRB14 |   | 24 | 62 | 179 | 181 | 0.113 | -0.129 | 0.143 | -0.098 | 0.138 |
|---|
| BCKDHA |   | 1 | 1195 | 1358 | 1387 | 0.091 | -0.045 | 0.072 | undef | 0.101 |
|---|
| ARHGEF7 |   | 25 | 59 | 83 | 92 | -0.074 | -0.061 | 0.208 | 0.028 | 0.015 |
|---|
| GABBR1 |   | 5 | 360 | 780 | 767 | -0.107 | 0.164 | 0.081 | 0.074 | 0.131 |
|---|
| PDK4 |   | 1 | 1195 | 1358 | 1387 | -0.027 | -0.030 | 0.216 | -0.130 | -0.041 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| SPSB1 |   | 3 | 557 | 1083 | 1073 | -0.124 | 0.088 | -0.152 | undef | 0.169 |
|---|
| ABL1 |   | 13 | 124 | 727 | 699 | -0.152 | 0.200 | 0.108 | undef | -0.059 |
|---|
| CCNA2 |   | 21 | 73 | 179 | 183 | 0.176 | -0.053 | 0.188 | -0.078 | 0.246 |
|---|
| CTGF |   | 32 | 40 | 318 | 290 | -0.169 | 0.181 | 0.126 | 0.101 | 0.209 |
|---|
| CDC42 |   | 26 | 52 | 179 | 177 | 0.047 | 0.242 | -0.027 | 0.040 | -0.042 |
|---|
| PAK2 |   | 26 | 52 | 179 | 177 | 0.014 | 0.161 | 0.081 | -0.044 | 0.062 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| TULP4 |   | 2 | 743 | 1358 | 1360 | -0.114 | 0.067 | 0.024 | -0.099 | 0.047 |
|---|
| GABBR2 |   | 1 | 1195 | 1358 | 1387 | 0.130 | 0.180 | 0.117 | 0.071 | 0.037 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| SKP2 |   | 29 | 45 | 1 | 23 | 0.092 | -0.043 | 0.237 | undef | -0.009 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| ZFPM2 |   | 5 | 360 | 525 | 518 | 0.001 | 0.092 | 0.113 | -0.099 | 0.379 |
|---|
| SKA1 |   | 2 | 743 | 1358 | 1360 | 0.081 | -0.085 | 0.037 | undef | -0.070 |
|---|
| PDHB |   | 13 | 124 | 525 | 503 | 0.056 | 0.017 | 0.197 | -0.205 | 0.193 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| NR2F1 |   | 25 | 59 | 83 | 92 | -0.252 | 0.113 | 0.133 | -0.116 | 0.207 |
|---|
| RASA1 |   | 2 | 743 | 1358 | 1360 | -0.079 | 0.146 | 0.127 | -0.056 | 0.149 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| HIF1A |   | 13 | 124 | 236 | 230 | -0.098 | 0.224 | -0.118 | 0.135 | 0.356 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| TIE1 |   | 18 | 86 | 366 | 357 | -0.108 | 0.005 | 0.141 | -0.108 | 0.252 |
|---|
| DSG1 |   | 10 | 167 | 658 | 636 | 0.072 | -0.075 | 0.252 | -0.061 | 0.068 |
|---|
| DLAT |   | 4 | 440 | 1141 | 1127 | 0.111 | 0.029 | 0.072 | undef | -0.014 |
|---|
GO Enrichment output for subnetwork 6808 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| branched chain family amino acid metabolic process | GO:0009081 |  | 1.931E-07 | 4.717E-04 |
|---|
| response to nutrient | GO:0007584 |  | 2.422E-07 | 2.959E-04 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 2.793E-07 | 2.274E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 4.81E-07 | 2.938E-04 |
|---|
| nuclear migration | GO:0007097 |  | 5.573E-07 | 2.723E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 5.573E-07 | 2.269E-04 |
|---|
| regulation of oxidoreductase activity | GO:0051341 |  | 8.48E-07 | 2.959E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 9.953E-07 | 3.039E-04 |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 1.187E-06 | 3.223E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.391E-06 | 3.397E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.553E-06 | 3.45E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| branched chain family amino acid metabolic process | GO:0009081 |  | 2.142E-07 | 5.154E-04 |
|---|
| response to nutrient | GO:0007584 |  | 2.423E-07 | 2.915E-04 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 3.027E-07 | 2.427E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 4.33E-07 | 2.604E-04 |
|---|
| nuclear migration | GO:0007097 |  | 6.039E-07 | 2.906E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 6.039E-07 | 2.422E-04 |
|---|
| regulation of oxidoreductase activity | GO:0051341 |  | 9.404E-07 | 3.232E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.033E-06 | 3.105E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.054E-06 | 2.819E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.054E-06 | 2.537E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.317E-06 | 2.88E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 5.112E-07 | 1.176E-03 |
|---|
| nuclear migration | GO:0007097 |  | 5.112E-07 | 5.879E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 5.112E-07 | 3.919E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.02E-06 | 5.863E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 1.02E-06 | 4.69E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.35E-06 | 5.176E-04 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 1.353E-06 | 4.446E-04 |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 1.554E-06 | 4.468E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.78E-06 | 4.548E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.78E-06 | 4.093E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.86E-06 | 3.889E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cell migration | GO:0030334 |  | 2.775E-09 | 5.114E-06 |
|---|
| regulation of locomotion | GO:0040012 |  | 5.944E-09 | 5.477E-06 |
|---|
| regulation of cell motion | GO:0051270 |  | 5.944E-09 | 3.652E-06 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.163E-07 | 5.358E-05 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.764E-07 | 6.502E-05 |
|---|
| regulation of anatomical structure morphogenesis | GO:0022603 |  | 3.595E-07 | 1.104E-04 |
|---|
| positive regulation of glucose metabolic process | GO:0010907 |  | 3.714E-07 | 9.777E-05 |
|---|
| regulation of cellular carbohydrate catabolic process | GO:0043471 |  | 7.41E-07 | 1.707E-04 |
|---|
| positive regulation of cellular catabolic process | GO:0031331 |  | 1.294E-06 | 2.649E-04 |
|---|
| positive regulation of carbohydrate metabolic process | GO:0045913 |  | 1.294E-06 | 2.384E-04 |
|---|
| regulation of generation of precursor metabolites and energy | GO:0043467 |  | 2.065E-06 | 3.46E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 5.112E-07 | 1.176E-03 |
|---|
| nuclear migration | GO:0007097 |  | 5.112E-07 | 5.879E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 5.112E-07 | 3.919E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.02E-06 | 5.863E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 1.02E-06 | 4.69E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.35E-06 | 5.176E-04 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 1.353E-06 | 4.446E-04 |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 1.554E-06 | 4.468E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.78E-06 | 4.548E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.78E-06 | 4.093E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.86E-06 | 3.889E-04 |
|---|



