Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6789
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7500 | 3.763e-03 | 6.240e-04 | 7.801e-01 | 1.832e-06 |
|---|
| Loi | 0.2308 | 7.949e-02 | 1.116e-02 | 4.263e-01 | 3.781e-04 |
|---|
| Schmidt | 0.6753 | 0.000e+00 | 0.000e+00 | 4.291e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7910 | 0.000e+00 | 0.000e+00 | 3.436e-03 | 0.000e+00 |
|---|
| Wang | 0.2634 | 2.446e-03 | 4.166e-02 | 3.861e-01 | 3.934e-05 |
|---|
Expression data for subnetwork 6789 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
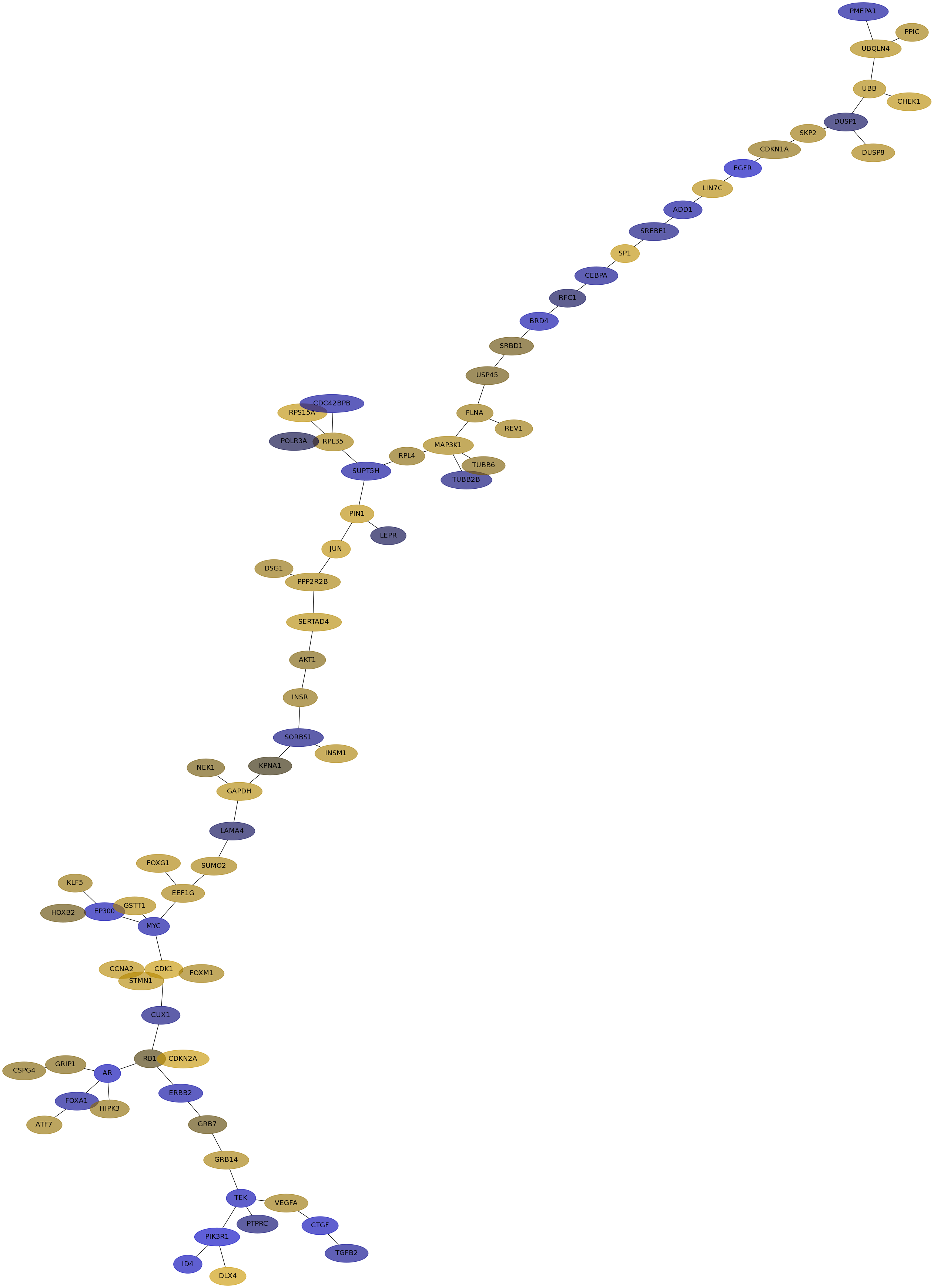
Subnetwork structure for each dataset
- IPC
Score for each gene in subnetwork 6789 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| TEK |   | 18 | 86 | 83 | 95 | -0.149 | -0.049 | 0.174 | 0.015 | 0.113 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| ADD1 |   | 8 | 222 | 366 | 373 | -0.085 | 0.116 | 0.035 | undef | -0.098 |
|---|
| DUSP8 |   | 4 | 440 | 1 | 59 | 0.118 | 0.257 | 0.068 | 0.208 | -0.029 |
|---|
| FOXM1 |   | 1 | 1195 | 1461 | 1479 | 0.103 | -0.204 | 0.051 | undef | -0.056 |
|---|
| SUMO2 |   | 22 | 69 | 366 | 345 | 0.117 | 0.062 | 0.294 | 0.141 | -0.096 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CEBPA |   | 3 | 557 | 1461 | 1440 | -0.058 | 0.005 | -0.100 | -0.199 | 0.039 |
|---|
| ID4 |   | 35 | 34 | 236 | 223 | -0.179 | 0.137 | 0.241 | undef | 0.036 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| TUBB6 |   | 3 | 557 | 1461 | 1440 | 0.045 | 0.114 | 0.187 | undef | 0.165 |
|---|
| EEF1G |   | 8 | 222 | 882 | 869 | 0.116 | -0.224 | 0.131 | 0.086 | 0.082 |
|---|
| STMN1 |   | 4 | 440 | 1461 | 1437 | 0.168 | -0.033 | 0.198 | 0.105 | 0.062 |
|---|
| CCNA2 |   | 21 | 73 | 179 | 183 | 0.176 | -0.053 | 0.188 | -0.078 | 0.246 |
|---|
| CTGF |   | 32 | 40 | 318 | 290 | -0.169 | 0.181 | 0.126 | 0.101 | 0.209 |
|---|
| PIN1 |   | 24 | 62 | 179 | 181 | 0.194 | 0.040 | -0.040 | 0.065 | -0.048 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| LAMA4 |   | 13 | 124 | 696 | 670 | -0.016 | 0.138 | 0.209 | 0.172 | 0.343 |
|---|
| UBQLN4 |   | 4 | 440 | 696 | 689 | 0.143 | -0.114 | undef | -0.213 | undef |
|---|
| GAPDH |   | 11 | 148 | 141 | 143 | 0.159 | -0.040 | -0.053 | 0.088 | 0.029 |
|---|
| RPL35 |   | 8 | 222 | 1323 | 1280 | 0.108 | -0.012 | 0.370 | -0.145 | -0.220 |
|---|
| BRD4 |   | 11 | 148 | 1047 | 1003 | -0.119 | 0.095 | -0.075 | 0.081 | -0.044 |
|---|
| AKT1 |   | 16 | 104 | 236 | 227 | 0.045 | 0.126 | -0.099 | undef | -0.168 |
|---|
| SERTAD4 |   | 8 | 222 | 1268 | 1225 | 0.173 | 0.169 | undef | -0.050 | undef |
|---|
| CUX1 |   | 6 | 301 | 1203 | 1169 | -0.042 | 0.256 | -0.121 | 0.105 | 0.040 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| USP45 |   | 1 | 1195 | 1461 | 1479 | 0.027 | 0.120 | undef | undef | undef |
|---|
| CDC42BPB |   | 1 | 1195 | 1461 | 1479 | -0.083 | 0.260 | 0.018 | 0.152 | -0.088 |
|---|
| DSG1 |   | 10 | 167 | 658 | 636 | 0.072 | -0.075 | 0.252 | -0.061 | 0.068 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| RFC1 |   | 1 | 1195 | 1461 | 1479 | -0.015 | 0.015 | 0.031 | -0.161 | -0.077 |
|---|
| GRB14 |   | 24 | 62 | 179 | 181 | 0.113 | -0.129 | 0.143 | -0.098 | 0.138 |
|---|
| ATF7 |   | 28 | 47 | 179 | 175 | 0.085 | 0.115 | -0.088 | 0.172 | -0.092 |
|---|
| GSTT1 |   | 15 | 111 | 1 | 32 | 0.148 | 0.107 | -0.190 | 0.022 | -0.064 |
|---|
| CHEK1 |   | 18 | 86 | 141 | 136 | 0.189 | -0.134 | 0.119 | -0.088 | -0.054 |
|---|
| TGFB2 |   | 7 | 256 | 525 | 515 | -0.063 | 0.244 | 0.226 | 0.079 | 0.015 |
|---|
| FOXG1 |   | 6 | 301 | 727 | 716 | 0.136 | -0.156 | -0.126 | -0.013 | -0.068 |
|---|
| HOXB2 |   | 33 | 37 | 318 | 289 | 0.024 | -0.143 | 0.076 | 0.247 | -0.184 |
|---|
| JUN |   | 26 | 52 | 179 | 177 | 0.200 | 0.051 | 0.220 | undef | 0.200 |
|---|
| RB1 |   | 31 | 43 | 1 | 22 | 0.013 | -0.051 | 0.057 | 0.100 | -0.082 |
|---|
| PPIC |   | 2 | 743 | 1461 | 1451 | 0.103 | 0.192 | -0.225 | 0.151 | 0.024 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| SREBF1 |   | 9 | 196 | 478 | 460 | -0.042 | 0.232 | -0.195 | 0.140 | -0.074 |
|---|
| GRIP1 |   | 3 | 557 | 696 | 695 | 0.043 | 0.104 | 0.000 | undef | undef |
|---|
| INSM1 |   | 23 | 66 | 570 | 545 | 0.135 | -0.184 | -0.188 | 0.237 | -0.032 |
|---|
| UBB |   | 3 | 557 | 696 | 695 | 0.146 | -0.027 | 0.012 | undef | -0.019 |
|---|
| DLX4 |   | 10 | 167 | 236 | 234 | 0.291 | 0.057 | -0.004 | undef | -0.059 |
|---|
| REV1 |   | 9 | 196 | 296 | 285 | 0.090 | -0.027 | 0.165 | 0.049 | 0.039 |
|---|
| DUSP1 |   | 4 | 440 | 1 | 59 | -0.019 | 0.127 | 0.170 | -0.031 | 0.134 |
|---|
| CSPG4 |   | 1 | 1195 | 1461 | 1479 | 0.051 | 0.088 | 0.190 | -0.096 | -0.027 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| MAP3K1 |   | 5 | 360 | 141 | 155 | 0.112 | 0.039 | undef | undef | undef |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| SKP2 |   | 29 | 45 | 1 | 23 | 0.092 | -0.043 | 0.237 | undef | -0.009 |
|---|
| SUPT5H |   | 6 | 301 | 1323 | 1281 | -0.085 | 0.007 | -0.298 | 0.002 | -0.068 |
|---|
| LIN7C |   | 10 | 167 | 590 | 566 | 0.166 | 0.088 | 0.118 | -0.018 | 0.094 |
|---|
| LEPR |   | 2 | 743 | 1461 | 1451 | -0.012 | 0.083 | -0.057 | -0.058 | 0.085 |
|---|
| POLR3A |   | 8 | 222 | 296 | 287 | -0.010 | -0.032 | undef | 0.091 | undef |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| PTPRC |   | 9 | 196 | 83 | 99 | -0.031 | -0.204 | -0.050 | -0.179 | -0.068 |
|---|
| KPNA1 |   | 9 | 196 | 141 | 147 | 0.007 | 0.152 | -0.083 | 0.073 | 0.086 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| NEK1 |   | 15 | 111 | 141 | 139 | 0.032 | 0.184 | 0.049 | undef | 0.135 |
|---|
| SRBD1 |   | 1 | 1195 | 1461 | 1479 | 0.025 | 0.045 | -0.258 | 0.138 | 0.291 |
|---|
| RPS15A |   | 2 | 743 | 842 | 854 | 0.221 | -0.019 | 0.189 | -0.123 | -0.060 |
|---|
| PMEPA1 |   | 3 | 557 | 1294 | 1276 | -0.085 | 0.137 | 0.167 | 0.087 | 0.102 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| TUBB2B |   | 13 | 124 | 141 | 141 | -0.036 | 0.304 | 0.045 | 0.187 | 0.139 |
|---|
| RPL4 |   | 1 | 1195 | 1461 | 1479 | 0.057 | 0.095 | 0.248 | -0.301 | -0.014 |
|---|
| HIPK3 |   | 9 | 196 | 1461 | 1417 | 0.069 | 0.005 | -0.027 | undef | 0.175 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
| FOXA1 |   | 27 | 50 | 179 | 176 | -0.072 | 0.071 | -0.015 | 0.214 | 0.005 |
|---|
GO Enrichment output for subnetwork 6789 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.135E-10 | 5.216E-07 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.675E-10 | 3.267E-07 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.058E-10 | 2.49E-07 |
|---|
| lung development | GO:0030324 |  | 6.115E-09 | 3.735E-06 |
|---|
| respiratory tube development | GO:0030323 |  | 8.627E-09 | 4.215E-06 |
|---|
| respiratory system development | GO:0060541 |  | 1.076E-08 | 4.38E-06 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 3.385E-08 | 1.181E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.709E-08 | 1.133E-05 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 8.363E-08 | 2.27E-05 |
|---|
| ear development | GO:0043583 |  | 1.046E-07 | 2.556E-05 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 1.407E-07 | 3.126E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.229E-10 | 1.018E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.509E-10 | 5.424E-07 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 6.052E-10 | 4.853E-07 |
|---|
| interphase | GO:0051325 |  | 6.149E-10 | 3.699E-07 |
|---|
| lung development | GO:0030324 |  | 9.455E-09 | 4.55E-06 |
|---|
| respiratory tube development | GO:0030323 |  | 1.347E-08 | 5.401E-06 |
|---|
| respiratory system development | GO:0060541 |  | 1.689E-08 | 5.807E-06 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 3.712E-08 | 1.116E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.707E-08 | 1.793E-05 |
|---|
| ear development | GO:0043583 |  | 1.215E-07 | 2.924E-05 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 1.291E-07 | 2.824E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.093E-09 | 4.813E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.869E-08 | 4.45E-05 |
|---|
| interphase | GO:0051325 |  | 4.74E-08 | 3.634E-05 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.141E-08 | 4.681E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 4.365E-07 | 2.008E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 4.93E-07 | 1.89E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 7.007E-07 | 2.302E-04 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 9.173E-07 | 2.637E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.093E-06 | 2.793E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.371E-06 | 3.154E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.703E-06 | 3.56E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| angiogenesis | GO:0001525 |  | 6.985E-07 | 1.287E-03 |
|---|
| blood vessel development | GO:0001568 |  | 7.455E-07 | 6.87E-04 |
|---|
| vasculature development | GO:0001944 |  | 8.77E-07 | 5.388E-04 |
|---|
| regulation of cell migration | GO:0030334 |  | 1.431E-06 | 6.594E-04 |
|---|
| regulation of locomotion | GO:0040012 |  | 2.967E-06 | 1.094E-03 |
|---|
| regulation of cell motion | GO:0051270 |  | 2.967E-06 | 9.114E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.048E-06 | 8.024E-04 |
|---|
| blood vessel morphogenesis | GO:0048514 |  | 4.145E-06 | 9.548E-04 |
|---|
| prostate gland development | GO:0030850 |  | 5.05E-06 | 1.034E-03 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 9.265E-06 | 1.707E-03 |
|---|
| regulation of embryonic development | GO:0045995 |  | 1.004E-05 | 1.683E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.093E-09 | 4.813E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.869E-08 | 4.45E-05 |
|---|
| interphase | GO:0051325 |  | 4.74E-08 | 3.634E-05 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 8.141E-08 | 4.681E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 4.365E-07 | 2.008E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 4.93E-07 | 1.89E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 7.007E-07 | 2.302E-04 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 9.173E-07 | 2.637E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.093E-06 | 2.793E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.371E-06 | 3.154E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.703E-06 | 3.56E-04 |
|---|



