Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6749
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7504 | 3.737e-03 | 6.190e-04 | 7.792e-01 | 1.802e-06 |
|---|
| Loi | 0.2310 | 7.926e-02 | 1.110e-02 | 4.258e-01 | 3.745e-04 |
|---|
| Schmidt | 0.6748 | 0.000e+00 | 0.000e+00 | 4.342e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7910 | 0.000e+00 | 0.000e+00 | 3.433e-03 | 0.000e+00 |
|---|
| Wang | 0.2632 | 2.471e-03 | 4.189e-02 | 3.872e-01 | 4.008e-05 |
|---|
Expression data for subnetwork 6749 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
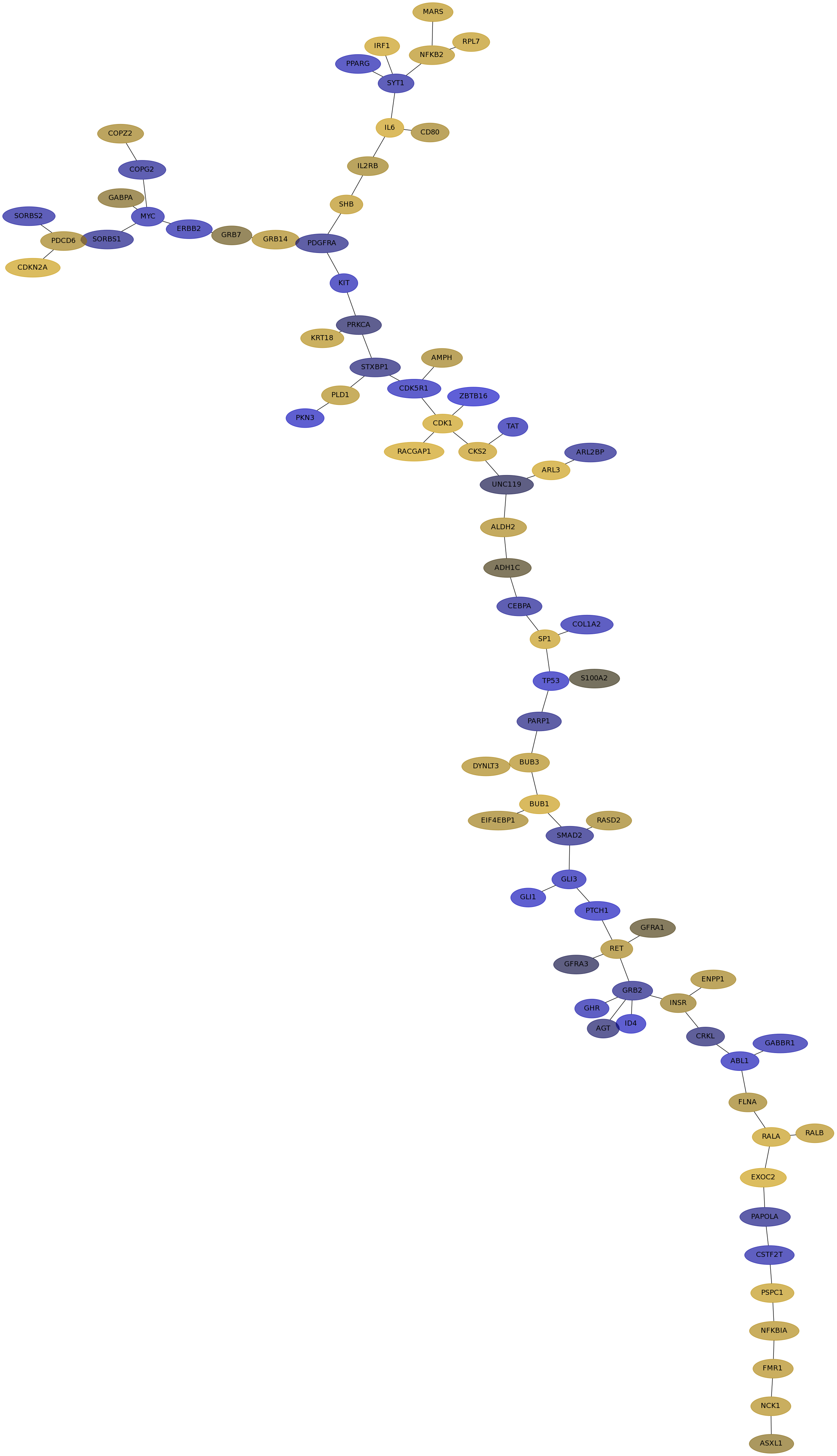
Score for each gene in subnetwork 6749 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| DYNLT3 |   | 2 | 743 | 590 | 616 | 0.117 | 0.208 | 0.030 | 0.137 | 0.187 |
|---|
| RET |   | 7 | 256 | 366 | 376 | 0.098 | 0.063 | -0.179 | undef | -0.159 |
|---|
| ARL2BP |   | 1 | 1195 | 1629 | 1645 | -0.048 | -0.023 | -0.181 | 0.170 | 0.168 |
|---|
| GFRA3 |   | 1 | 1195 | 1629 | 1645 | -0.009 | -0.112 | -0.120 | -0.207 | 0.012 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| ID4 |   | 35 | 34 | 236 | 223 | -0.179 | 0.137 | 0.241 | undef | 0.036 |
|---|
| CEBPA |   | 3 | 557 | 1461 | 1440 | -0.058 | 0.005 | -0.100 | -0.199 | 0.039 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| PDGFRA |   | 10 | 167 | 141 | 146 | -0.035 | 0.008 | 0.118 | 0.107 | 0.119 |
|---|
| PAPOLA |   | 1 | 1195 | 1629 | 1645 | -0.042 | -0.072 | 0.179 | 0.092 | 0.168 |
|---|
| CRKL |   | 6 | 301 | 780 | 764 | -0.023 | 0.056 | 0.271 | -0.025 | -0.008 |
|---|
| RALB |   | 2 | 743 | 1474 | 1462 | 0.150 | 0.137 | -0.036 | -0.047 | 0.007 |
|---|
| EXOC2 |   | 2 | 743 | 1558 | 1545 | 0.279 | 0.060 | -0.105 | 0.067 | -0.098 |
|---|
| NFKB2 |   | 7 | 256 | 1116 | 1082 | 0.153 | -0.239 | -0.131 | undef | -0.065 |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| CKS2 |   | 9 | 196 | 928 | 903 | 0.214 | -0.220 | 0.089 | -0.003 | -0.056 |
|---|
| GLI1 |   | 1 | 1195 | 1629 | 1645 | -0.172 | 0.028 | -0.131 | 0.225 | 0.125 |
|---|
| TAT |   | 11 | 148 | 83 | 97 | -0.113 | 0.030 | -0.182 | 0.037 | -0.074 |
|---|
| ZBTB16 |   | 1 | 1195 | 1629 | 1645 | -0.232 | 0.113 | -0.028 | -0.051 | -0.067 |
|---|
| PARP1 |   | 29 | 45 | 83 | 84 | -0.036 | -0.162 | -0.089 | 0.238 | 0.012 |
|---|
| PPARG |   | 7 | 256 | 179 | 201 | -0.119 | 0.011 | 0.105 | 0.059 | 0.039 |
|---|
| GABPA |   | 23 | 66 | 696 | 665 | 0.033 | -0.183 | 0.066 | 0.141 | -0.161 |
|---|
| NCK1 |   | 3 | 557 | 318 | 341 | 0.135 | -0.100 | -0.059 | -0.108 | 0.151 |
|---|
| ASXL1 |   | 2 | 743 | 1510 | 1502 | 0.044 | 0.146 | 0.031 | undef | 0.041 |
|---|
| PTCH1 |   | 5 | 360 | 1190 | 1157 | -0.184 | 0.047 | 0.121 | -0.055 | 0.004 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| RALA |   | 10 | 167 | 318 | 323 | 0.224 | 0.207 | -0.047 | 0.169 | 0.146 |
|---|
| ENPP1 |   | 5 | 360 | 1055 | 1034 | 0.088 | 0.129 | -0.026 | undef | 0.039 |
|---|
| SMAD2 |   | 4 | 440 | 590 | 585 | -0.039 | -0.199 | 0.076 | -0.003 | 0.247 |
|---|
| EIF4EBP1 |   | 3 | 557 | 1222 | 1210 | 0.086 | -0.201 | -0.147 | 0.183 | 0.216 |
|---|
| SHB |   | 2 | 743 | 236 | 272 | 0.164 | 0.141 | -0.074 | 0.107 | -0.048 |
|---|
| COPZ2 |   | 2 | 743 | 1629 | 1614 | 0.081 | 0.013 | 0.127 | 0.126 | 0.123 |
|---|
| GRB14 |   | 24 | 62 | 179 | 181 | 0.113 | -0.129 | 0.143 | -0.098 | 0.138 |
|---|
| GABBR1 |   | 5 | 360 | 780 | 767 | -0.107 | 0.164 | 0.081 | 0.074 | 0.131 |
|---|
| CSTF2T |   | 1 | 1195 | 1629 | 1645 | -0.102 | -0.245 | 0.140 | 0.074 | 0.109 |
|---|
| RACGAP1 |   | 4 | 440 | 1222 | 1205 | 0.283 | -0.164 | 0.073 | 0.055 | 0.188 |
|---|
| S100A2 |   | 6 | 301 | 1 | 52 | 0.005 | 0.047 | 0.137 | -0.029 | -0.008 |
|---|
| COPG2 |   | 4 | 440 | 412 | 419 | -0.058 | -0.232 | 0.000 | 0.086 | -0.081 |
|---|
| ARL3 |   | 6 | 301 | 83 | 105 | 0.266 | -0.002 | 0.203 | 0.206 | -0.029 |
|---|
| ABL1 |   | 13 | 124 | 727 | 699 | -0.152 | 0.200 | 0.108 | undef | -0.059 |
|---|
| UNC119 |   | 10 | 167 | 83 | 98 | -0.009 | -0.324 | -0.099 | 0.094 | 0.071 |
|---|
| GLI3 |   | 21 | 73 | 478 | 456 | -0.138 | 0.080 | -0.007 | 0.282 | -0.018 |
|---|
| AMPH |   | 7 | 256 | 83 | 102 | 0.082 | 0.000 | -0.104 | 0.024 | 0.241 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| RASD2 |   | 1 | 1195 | 1629 | 1645 | 0.085 | -0.069 | undef | -0.008 | undef |
|---|
| BUB1 |   | 6 | 301 | 590 | 582 | 0.246 | -0.172 | 0.013 | -0.007 | 0.170 |
|---|
| STXBP1 |   | 5 | 360 | 1629 | 1587 | -0.027 | 0.078 | 0.180 | 0.143 | 0.043 |
|---|
| FMR1 |   | 2 | 743 | 1604 | 1588 | 0.137 | -0.104 | 0.352 | -0.014 | 0.113 |
|---|
| GFRA1 |   | 2 | 743 | 366 | 395 | 0.011 | -0.067 | 0.292 | 0.051 | 0.029 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| ALDH2 |   | 1 | 1195 | 1629 | 1645 | 0.111 | -0.121 | 0.002 | 0.114 | 0.105 |
|---|
| BUB3 |   | 5 | 360 | 478 | 476 | 0.133 | -0.053 | 0.165 | 0.195 | 0.181 |
|---|
| PSPC1 |   | 6 | 301 | 991 | 964 | 0.203 | -0.289 | 0.298 | 0.148 | -0.107 |
|---|
| RPL7 |   | 3 | 557 | 1437 | 1418 | 0.189 | -0.121 | 0.166 | 0.071 | -0.003 |
|---|
| CD80 |   | 4 | 440 | 1055 | 1035 | 0.081 | -0.199 | -0.136 | 0.034 | -0.078 |
|---|
| MARS |   | 2 | 743 | 1364 | 1363 | 0.169 | 0.024 | 0.010 | 0.156 | 0.060 |
|---|
| SORBS2 |   | 1 | 1195 | 1629 | 1645 | -0.075 | 0.077 | 0.084 | -0.038 | -0.114 |
|---|
| IL2RB |   | 1 | 1195 | 1629 | 1645 | 0.079 | -0.240 | -0.175 | -0.177 | -0.068 |
|---|
| SYT1 |   | 11 | 148 | 1 | 39 | -0.079 | 0.051 | 0.038 | 0.189 | -0.124 |
|---|
| CDK5R1 |   | 17 | 95 | 552 | 521 | -0.150 | 0.038 | -0.096 | 0.121 | 0.091 |
|---|
| ADH1C |   | 2 | 743 | 1629 | 1614 | 0.009 | 0.062 | 0.163 | 0.059 | -0.024 |
|---|
| GRB2 |   | 13 | 124 | 318 | 316 | -0.040 | -0.159 | -0.036 | -0.028 | -0.024 |
|---|
| PDCD6 |   | 10 | 167 | 1 | 40 | 0.091 | -0.060 | undef | undef | undef |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| AGT |   | 4 | 440 | 1203 | 1184 | -0.020 | -0.101 | -0.043 | 0.255 | 0.064 |
|---|
| PKN3 |   | 18 | 86 | 179 | 187 | -0.181 | 0.009 | undef | 0.239 | undef |
|---|
| IL6 |   | 9 | 196 | 366 | 365 | 0.257 | -0.106 | 0.226 | 0.024 | 0.035 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| NFKBIA |   | 1 | 1195 | 1629 | 1645 | 0.133 | -0.146 | 0.007 | undef | -0.334 |
|---|
| PLD1 |   | 17 | 95 | 1 | 29 | 0.132 | 0.033 | -0.158 | -0.062 | 0.123 |
|---|
| COL1A2 |   | 7 | 256 | 552 | 542 | -0.110 | 0.207 | 0.046 | 0.235 | 0.097 |
|---|
| KRT18 |   | 8 | 222 | 590 | 569 | 0.148 | 0.100 | -0.083 | 0.024 | -0.053 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
| IRF1 |   | 3 | 557 | 1629 | 1610 | 0.241 | -0.216 | -0.131 | -0.037 | -0.170 |
|---|
GO Enrichment output for subnetwork 6749 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 5.811E-09 | 1.42E-05 |
|---|
| peptidyl-tyrosine modification | GO:0018212 |  | 9.421E-09 | 1.151E-05 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 3.227E-08 | 2.628E-05 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 6.591E-08 | 4.025E-05 |
|---|
| positive regulation of inflammatory response | GO:0050729 |  | 9.651E-08 | 4.715E-05 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 1.206E-07 | 4.909E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.883E-07 | 6.57E-05 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 1.911E-07 | 5.836E-05 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 1.964E-07 | 5.332E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.989E-07 | 4.858E-05 |
|---|
| response to insulin stimulus | GO:0032868 |  | 2.459E-07 | 5.461E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 7.903E-09 | 1.902E-05 |
|---|
| peptidyl-tyrosine modification | GO:0018212 |  | 1.281E-08 | 1.541E-05 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 3.981E-08 | 3.193E-05 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 7.564E-08 | 4.55E-05 |
|---|
| positive regulation of inflammatory response | GO:0050729 |  | 1.249E-07 | 6.011E-05 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 1.389E-07 | 5.571E-05 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 2.296E-07 | 7.891E-05 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 2.356E-07 | 7.085E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.549E-07 | 6.814E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.572E-07 | 6.189E-05 |
|---|
| response to insulin stimulus | GO:0032868 |  | 2.917E-07 | 6.38E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 1.248E-08 | 2.871E-05 |
|---|
| peptidyl-tyrosine modification | GO:0018212 |  | 2.164E-08 | 2.488E-05 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 9.435E-08 | 7.233E-05 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.557E-07 | 8.952E-05 |
|---|
| positive regulation of inflammatory response | GO:0050729 |  | 2.762E-07 | 1.27E-04 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 3.358E-07 | 1.287E-04 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 3.764E-07 | 1.237E-04 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 4.201E-07 | 1.208E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.286E-07 | 1.095E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 5.889E-07 | 1.354E-04 |
|---|
| response to insulin stimulus | GO:0032868 |  | 6.687E-07 | 1.398E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 4.903E-07 | 9.036E-04 |
|---|
| peptidyl-tyrosine modification | GO:0018212 |  | 6.493E-07 | 5.983E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 8.464E-07 | 5.2E-04 |
|---|
| cytokine-mediated signaling pathway | GO:0019221 |  | 9.822E-07 | 4.526E-04 |
|---|
| Ras protein signal transduction | GO:0007265 |  | 1.737E-06 | 6.404E-04 |
|---|
| response to hormone stimulus | GO:0009725 |  | 2.344E-06 | 7.201E-04 |
|---|
| response to insulin stimulus | GO:0032868 |  | 2.645E-06 | 6.963E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 2.687E-06 | 6.191E-04 |
|---|
| actin cytoskeleton reorganization | GO:0031532 |  | 6.55E-06 | 1.341E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 7.098E-06 | 1.308E-03 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 1.059E-05 | 1.775E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 1.248E-08 | 2.871E-05 |
|---|
| peptidyl-tyrosine modification | GO:0018212 |  | 2.164E-08 | 2.488E-05 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 9.435E-08 | 7.233E-05 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.557E-07 | 8.952E-05 |
|---|
| positive regulation of inflammatory response | GO:0050729 |  | 2.762E-07 | 1.27E-04 |
|---|
| regulation of insulin receptor signaling pathway | GO:0046626 |  | 3.358E-07 | 1.287E-04 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 3.764E-07 | 1.237E-04 |
|---|
| positive regulation of protein amino acid phosphorylation | GO:0001934 |  | 4.201E-07 | 1.208E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.286E-07 | 1.095E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 5.889E-07 | 1.354E-04 |
|---|
| response to insulin stimulus | GO:0032868 |  | 6.687E-07 | 1.398E-04 |
|---|



