Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6709
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7513 | 3.680e-03 | 6.070e-04 | 7.771e-01 | 1.736e-06 |
|---|
| Loi | 0.2312 | 7.889e-02 | 1.100e-02 | 4.249e-01 | 3.687e-04 |
|---|
| Schmidt | 0.6745 | 0.000e+00 | 0.000e+00 | 4.376e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7909 | 0.000e+00 | 0.000e+00 | 3.450e-03 | 0.000e+00 |
|---|
| Wang | 0.2629 | 2.509e-03 | 4.225e-02 | 3.890e-01 | 4.124e-05 |
|---|
Expression data for subnetwork 6709 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
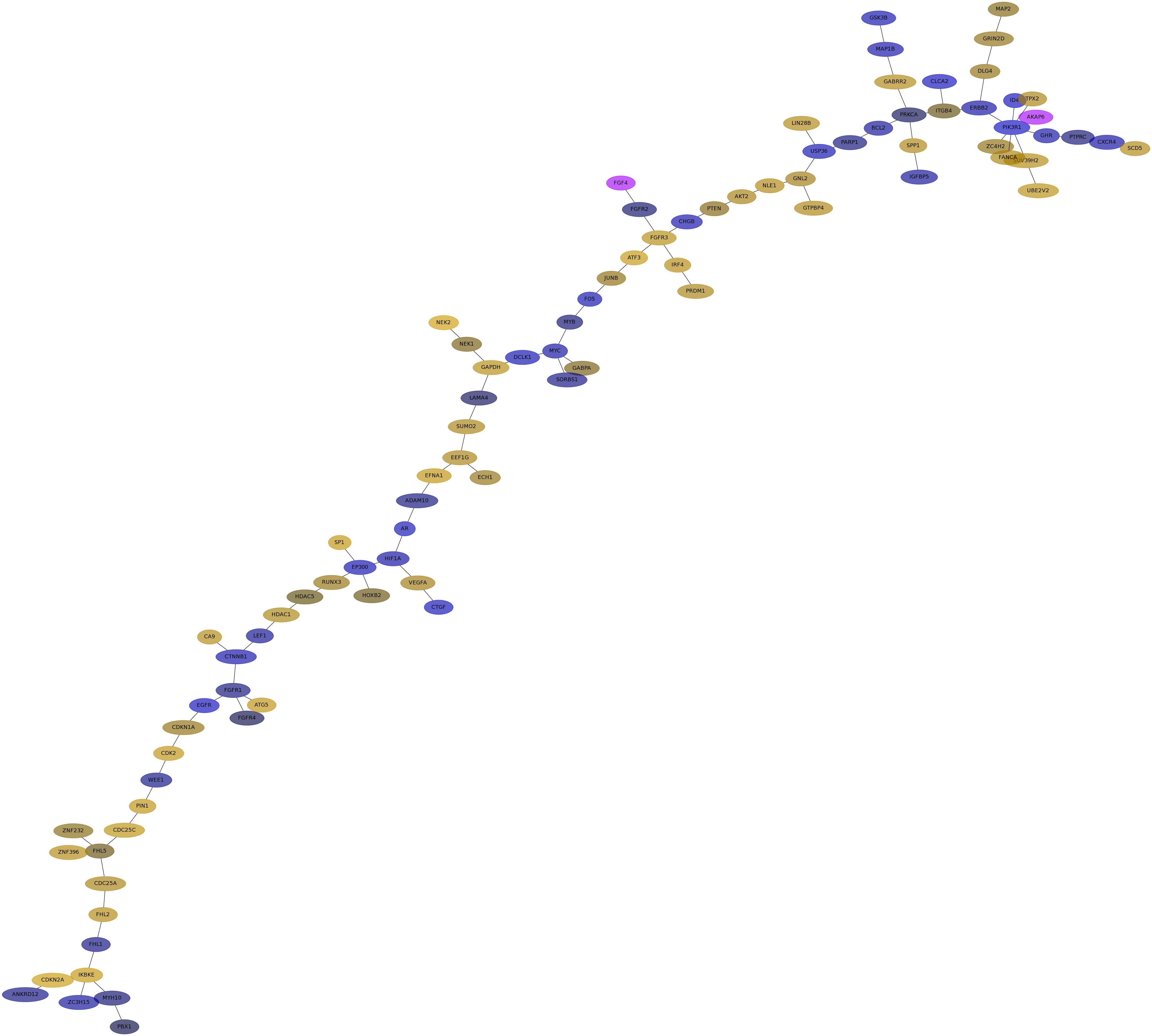
Score for each gene in subnetwork 6709 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| LIN28B |   | 1 | 1195 | 882 | 918 | 0.136 | 0.071 | undef | undef | undef |
|---|
| ADAM10 |   | 1 | 1195 | 882 | 918 | -0.036 | 0.069 | 0.151 | -0.107 | -0.086 |
|---|
| SUMO2 |   | 22 | 69 | 366 | 345 | 0.117 | 0.062 | 0.294 | 0.141 | -0.096 |
|---|
| TPX2 |   | 5 | 360 | 412 | 416 | 0.114 | -0.121 | -0.018 | undef | -0.056 |
|---|
| PBX1 |   | 6 | 301 | 1 | 52 | -0.010 | 0.145 | -0.013 | 0.176 | -0.053 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| ID4 |   | 35 | 34 | 236 | 223 | -0.179 | 0.137 | 0.241 | undef | 0.036 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| NLE1 |   | 3 | 557 | 882 | 871 | 0.138 | -0.086 | -0.018 | 0.001 | 0.044 |
|---|
| EEF1G |   | 8 | 222 | 882 | 869 | 0.116 | -0.224 | 0.131 | 0.086 | 0.082 |
|---|
| MYH10 |   | 5 | 360 | 236 | 242 | -0.029 | 0.139 | 0.132 | undef | 0.223 |
|---|
| WEE1 |   | 4 | 440 | 658 | 649 | -0.051 | 0.163 | 0.034 | undef | 0.215 |
|---|
| ZC3H15 |   | 1 | 1195 | 882 | 918 | -0.081 | 0.097 | -0.015 | 0.030 | -0.030 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| CTGF |   | 32 | 40 | 318 | 290 | -0.169 | 0.181 | 0.126 | 0.101 | 0.209 |
|---|
| PIN1 |   | 24 | 62 | 179 | 181 | 0.194 | 0.040 | -0.040 | 0.065 | -0.048 |
|---|
| ANKRD12 |   | 1 | 1195 | 882 | 918 | -0.047 | -0.193 | 0.074 | -0.122 | 0.094 |
|---|
| LAMA4 |   | 13 | 124 | 696 | 670 | -0.016 | 0.138 | 0.209 | 0.172 | 0.343 |
|---|
| GAPDH |   | 11 | 148 | 141 | 143 | 0.159 | -0.040 | -0.053 | 0.088 | 0.029 |
|---|
| PARP1 |   | 29 | 45 | 83 | 84 | -0.036 | -0.162 | -0.089 | 0.238 | 0.012 |
|---|
| FGF4 |   | 2 | 743 | 882 | 892 | undef | undef | undef | 0.116 | undef |
|---|
| GABPA |   | 23 | 66 | 696 | 665 | 0.033 | -0.183 | 0.066 | 0.141 | -0.161 |
|---|
| AKT2 |   | 28 | 47 | 412 | 394 | 0.110 | -0.118 | -0.003 | 0.088 | -0.092 |
|---|
| DLG4 |   | 5 | 360 | 806 | 784 | 0.059 | -0.113 | 0.151 | -0.131 | 0.123 |
|---|
| ATF3 |   | 17 | 95 | 179 | 189 | 0.225 | -0.021 | 0.000 | undef | -0.039 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| CDC25C |   | 3 | 557 | 658 | 654 | 0.189 | 0.029 | 0.201 | -0.055 | -0.062 |
|---|
| HDAC5 |   | 2 | 743 | 882 | 892 | 0.022 | 0.029 | -0.165 | -0.125 | 0.009 |
|---|
| FGFR1 |   | 26 | 52 | 638 | 615 | -0.036 | 0.200 | 0.087 | undef | -0.015 |
|---|
| USP36 |   | 3 | 557 | 882 | 871 | -0.144 | -0.044 | 0.203 | undef | 0.014 |
|---|
| ZNF396 |   | 2 | 743 | 882 | 892 | 0.151 | undef | undef | undef | undef |
|---|
| ZNF232 |   | 3 | 557 | 882 | 871 | 0.048 | -0.040 | 0.189 | -0.007 | 0.159 |
|---|
| LEF1 |   | 3 | 557 | 696 | 695 | -0.068 | 0.016 | -0.018 | 0.001 | 0.139 |
|---|
| DCLK1 |   | 7 | 256 | 141 | 151 | -0.145 | -0.006 | -0.048 | 0.188 | -0.035 |
|---|
| UBE2V2 |   | 2 | 743 | 1 | 66 | 0.180 | -0.027 | 0.196 | undef | 0.060 |
|---|
| MAP2 |   | 3 | 557 | 552 | 558 | 0.044 | 0.045 | 0.014 | undef | -0.057 |
|---|
| CLCA2 |   | 9 | 196 | 412 | 411 | -0.176 | 0.243 | -0.153 | 0.191 | -0.000 |
|---|
| HDAC1 |   | 4 | 440 | 318 | 337 | 0.122 | -0.020 | 0.050 | -0.035 | -0.172 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| ATG5 |   | 2 | 743 | 882 | 892 | 0.195 | -0.113 | -0.013 | -0.050 | 0.210 |
|---|
| CA9 |   | 2 | 743 | 882 | 892 | 0.146 | 0.137 | 0.068 | 0.099 | 0.116 |
|---|
| SCD5 |   | 1 | 1195 | 882 | 918 | 0.136 | 0.036 | 0.171 | -0.111 | -0.232 |
|---|
| GNL2 |   | 2 | 743 | 882 | 892 | 0.092 | 0.129 | -0.037 | -0.220 | -0.019 |
|---|
| GABRR2 |   | 3 | 557 | 882 | 871 | 0.135 | -0.008 | 0.204 | 0.225 | 0.115 |
|---|
| HOXB2 |   | 33 | 37 | 318 | 289 | 0.024 | -0.143 | 0.076 | 0.247 | -0.184 |
|---|
| AKAP6 |   | 3 | 557 | 882 | 871 | undef | 0.116 | -0.125 | 0.026 | 0.153 |
|---|
| IRF4 |   | 2 | 743 | 882 | 892 | 0.153 | -0.250 | -0.172 | -0.076 | -0.103 |
|---|
| FGFR3 |   | 9 | 196 | 179 | 195 | 0.160 | 0.071 | 0.018 | -0.050 | -0.111 |
|---|
| SPP1 |   | 26 | 52 | 83 | 91 | 0.130 | 0.119 | 0.184 | 0.204 | 0.153 |
|---|
| SUV39H2 |   | 8 | 222 | 318 | 325 | 0.150 | -0.097 | -0.057 | undef | 0.028 |
|---|
| PTEN |   | 27 | 50 | 141 | 135 | 0.045 | 0.055 | 0.017 | 0.183 | -0.041 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| JUNB |   | 20 | 81 | 179 | 185 | 0.057 | 0.086 | 0.065 | 0.001 | -0.015 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| NEK2 |   | 8 | 222 | 141 | 149 | 0.292 | -0.106 | 0.020 | 0.083 | 0.165 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| MYB |   | 28 | 47 | 366 | 344 | -0.028 | 0.075 | 0.002 | 0.065 | -0.012 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| CHGB |   | 7 | 256 | 682 | 664 | -0.126 | -0.041 | 0.018 | 0.102 | -0.131 |
|---|
| FGFR4 |   | 21 | 73 | 179 | 183 | -0.011 | 0.188 | -0.089 | -0.018 | -0.049 |
|---|
| CDC25A |   | 8 | 222 | 570 | 555 | 0.113 | -0.170 | 0.151 | -0.301 | -0.003 |
|---|
| ZC4H2 |   | 17 | 95 | 570 | 546 | 0.065 | 0.061 | 0.304 | -0.072 | 0.059 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| GTPBP4 |   | 4 | 440 | 882 | 870 | 0.129 | -0.179 | 0.076 | -0.043 | -0.026 |
|---|
| FHL2 |   | 21 | 73 | 658 | 630 | 0.143 | 0.239 | -0.033 | 0.107 | 0.294 |
|---|
| RUNX3 |   | 1 | 1195 | 882 | 918 | 0.071 | -0.097 | 0.083 | -0.106 | -0.074 |
|---|
| PRDM1 |   | 2 | 743 | 882 | 892 | 0.114 | -0.101 | undef | -0.251 | undef |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| FANCA |   | 9 | 196 | 1 | 44 | 0.120 | -0.188 | 0.106 | -0.127 | -0.053 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| ECH1 |   | 2 | 743 | 882 | 892 | 0.065 | -0.008 | -0.070 | 0.193 | -0.017 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| PTPRC |   | 9 | 196 | 83 | 99 | -0.031 | -0.204 | -0.050 | -0.179 | -0.068 |
|---|
| NEK1 |   | 15 | 111 | 141 | 139 | 0.032 | 0.184 | 0.049 | undef | 0.135 |
|---|
| CXCR4 |   | 2 | 743 | 882 | 892 | -0.103 | -0.189 | -0.094 | -0.160 | -0.010 |
|---|
| IKBKE |   | 10 | 167 | 552 | 539 | 0.227 | -0.041 | -0.095 | -0.197 | 0.079 |
|---|
| HIF1A |   | 13 | 124 | 236 | 230 | -0.098 | 0.224 | -0.118 | 0.135 | 0.356 |
|---|
| FGFR2 |   | 7 | 256 | 179 | 201 | -0.024 | -0.001 | 0.129 | undef | 0.009 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| GRIN2D |   | 13 | 124 | 552 | 522 | 0.058 | 0.155 | 0.000 | undef | undef |
|---|
| EFNA1 |   | 1 | 1195 | 882 | 918 | 0.198 | 0.099 | 0.139 | 0.002 | 0.082 |
|---|
| FHL1 |   | 14 | 117 | 614 | 591 | -0.041 | 0.104 | 0.301 | undef | 0.266 |
|---|
| ITGB4 |   | 35 | 34 | 1 | 20 | 0.021 | 0.313 | -0.134 | 0.080 | -0.114 |
|---|
| CDK2 |   | 31 | 43 | 179 | 174 | 0.196 | -0.096 | 0.109 | 0.001 | 0.362 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
| FHL5 |   | 3 | 557 | 658 | 654 | 0.022 | 0.031 | 0.145 | -0.095 | 0.153 |
|---|
GO Enrichment output for subnetwork 6709 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.05E-11 | 2.565E-08 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.521E-11 | 3.08E-08 |
|---|
| regulation of cell adhesion | GO:0030155 |  | 1.405E-09 | 1.144E-06 |
|---|
| negative regulation of cell adhesion | GO:0007162 |  | 8.146E-09 | 4.975E-06 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 8.475E-09 | 4.141E-06 |
|---|
| regulation of protein stability | GO:0031647 |  | 9.924E-09 | 4.041E-06 |
|---|
| regulation of cell-matrix adhesion | GO:0001952 |  | 1.227E-08 | 4.283E-06 |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 1.328E-08 | 4.054E-06 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.627E-08 | 4.415E-06 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 2.749E-08 | 6.715E-06 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 3.828E-08 | 8.501E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.826E-11 | 4.394E-08 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 6.094E-10 | 7.331E-07 |
|---|
| regulation of cell adhesion | GO:0030155 |  | 1.737E-09 | 1.393E-06 |
|---|
| negative regulation of cell adhesion | GO:0007162 |  | 7.987E-09 | 4.804E-06 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 1.131E-08 | 5.443E-06 |
|---|
| regulation of protein stability | GO:0031647 |  | 1.207E-08 | 4.84E-06 |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 1.676E-08 | 5.761E-06 |
|---|
| regulation of cell-matrix adhesion | GO:0001952 |  | 1.727E-08 | 5.194E-06 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.288E-08 | 6.116E-06 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 3.864E-08 | 9.296E-06 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 4.688E-08 | 1.025E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.831E-09 | 4.211E-06 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.786E-09 | 3.204E-06 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 8.372E-08 | 6.419E-05 |
|---|
| cell growth | GO:0016049 |  | 1.09E-07 | 6.269E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.444E-07 | 6.645E-05 |
|---|
| regulation of cell adhesion | GO:0030155 |  | 1.501E-07 | 5.755E-05 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 2.174E-07 | 7.144E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 6.734E-07 | 1.936E-04 |
|---|
| regulation of protein stability | GO:0031647 |  | 6.971E-07 | 1.782E-04 |
|---|
| negative regulation of cell adhesion | GO:0007162 |  | 8.45E-07 | 1.943E-04 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 1.34E-06 | 2.802E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cell migration | GO:0030334 |  | 2.124E-11 | 3.915E-08 |
|---|
| regulation of locomotion | GO:0040012 |  | 7.096E-11 | 6.539E-08 |
|---|
| regulation of cell motion | GO:0051270 |  | 7.096E-11 | 4.359E-08 |
|---|
| regulation of cell adhesion | GO:0030155 |  | 4.934E-07 | 2.274E-04 |
|---|
| placenta development | GO:0001890 |  | 5.324E-07 | 1.962E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 6.603E-07 | 2.028E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.083E-06 | 2.851E-04 |
|---|
| blood vessel development | GO:0001568 |  | 1.659E-06 | 3.823E-04 |
|---|
| vasculature development | GO:0001944 |  | 1.949E-06 | 3.991E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.804E-06 | 5.169E-04 |
|---|
| negative regulation of cell migration | GO:0030336 |  | 2.869E-06 | 4.807E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.831E-09 | 4.211E-06 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.786E-09 | 3.204E-06 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 8.372E-08 | 6.419E-05 |
|---|
| cell growth | GO:0016049 |  | 1.09E-07 | 6.269E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.444E-07 | 6.645E-05 |
|---|
| regulation of cell adhesion | GO:0030155 |  | 1.501E-07 | 5.755E-05 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 2.174E-07 | 7.144E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 6.734E-07 | 1.936E-04 |
|---|
| regulation of protein stability | GO:0031647 |  | 6.971E-07 | 1.782E-04 |
|---|
| negative regulation of cell adhesion | GO:0007162 |  | 8.45E-07 | 1.943E-04 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 1.34E-06 | 2.802E-04 |
|---|



