Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6695
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7516 | 3.661e-03 | 6.030e-04 | 7.764e-01 | 1.714e-06 |
|---|
| Loi | 0.2312 | 7.894e-02 | 1.101e-02 | 4.250e-01 | 3.695e-04 |
|---|
| Schmidt | 0.6743 | 0.000e+00 | 0.000e+00 | 4.392e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7909 | 0.000e+00 | 0.000e+00 | 3.448e-03 | 0.000e+00 |
|---|
| Wang | 0.2627 | 2.528e-03 | 4.243e-02 | 3.898e-01 | 4.181e-05 |
|---|
Expression data for subnetwork 6695 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
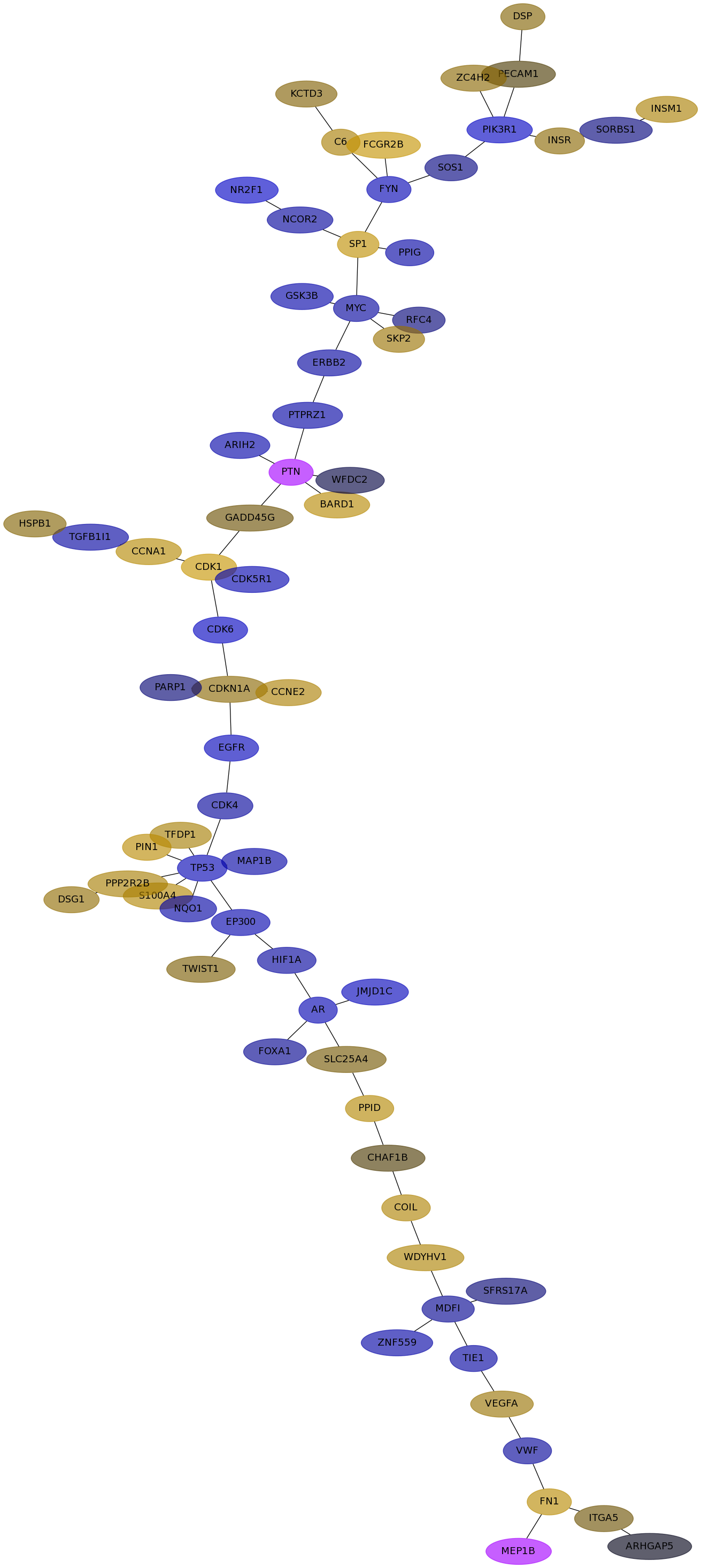
Score for each gene in subnetwork 6695 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| CCNE2 |   | 11 | 148 | 412 | 407 | 0.136 | -0.178 | 0.241 | undef | -0.055 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| NQO1 |   | 21 | 73 | 366 | 354 | -0.115 | 0.069 | -0.189 | 0.321 | 0.118 |
|---|
| WDYHV1 |   | 1 | 1195 | 913 | 953 | 0.144 | -0.125 | 0.015 | 0.113 | -0.054 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| WFDC2 |   | 5 | 360 | 236 | 242 | -0.011 | -0.011 | 0.333 | -0.162 | -0.157 |
|---|
| PPID |   | 1 | 1195 | 913 | 953 | 0.167 | -0.051 | -0.058 | 0.028 | -0.078 |
|---|
| PIN1 |   | 24 | 62 | 179 | 181 | 0.194 | 0.040 | -0.040 | 0.065 | -0.048 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| CDK6 |   | 16 | 104 | 318 | 314 | -0.229 | 0.107 | -0.028 | -0.118 | 0.104 |
|---|
| RFC4 |   | 1 | 1195 | 913 | 953 | -0.041 | 0.039 | 0.131 | 0.021 | 0.004 |
|---|
| ZNF559 |   | 3 | 557 | 913 | 908 | -0.122 | 0.089 | 0.145 | -0.005 | -0.014 |
|---|
| PARP1 |   | 29 | 45 | 83 | 84 | -0.036 | -0.162 | -0.089 | 0.238 | 0.012 |
|---|
| S100A4 |   | 2 | 743 | 913 | 914 | 0.163 | -0.116 | 0.096 | -0.146 | 0.148 |
|---|
| CDK4 |   | 4 | 440 | 913 | 904 | -0.092 | -0.201 | 0.091 | -0.043 | -0.017 |
|---|
| ARHGAP5 |   | 1 | 1195 | 913 | 953 | -0.003 | 0.321 | 0.083 | 0.047 | -0.005 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| DSP |   | 7 | 256 | 590 | 570 | 0.054 | 0.235 | -0.119 | -0.033 | 0.053 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| FYN |   | 10 | 167 | 1 | 40 | -0.171 | -0.167 | 0.073 | -0.137 | 0.110 |
|---|
| SLC25A4 |   | 2 | 743 | 913 | 914 | 0.038 | 0.267 | -0.117 | 0.158 | 0.141 |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| DSG1 |   | 10 | 167 | 658 | 636 | 0.072 | -0.075 | 0.252 | -0.061 | 0.068 |
|---|
| HSPB1 |   | 22 | 69 | 1 | 25 | 0.053 | 0.169 | 0.017 | 0.179 | 0.161 |
|---|
| COIL |   | 4 | 440 | 913 | 904 | 0.151 | 0.144 | 0.071 | 0.106 | 0.044 |
|---|
| PTN |   | 24 | 62 | 236 | 225 | undef | 0.022 | 0.000 | 0.054 | -0.092 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| NCOR2 |   | 32 | 40 | 318 | 290 | -0.096 | 0.211 | -0.163 | 0.158 | 0.114 |
|---|
| FN1 |   | 6 | 301 | 590 | 582 | 0.190 | 0.133 | 0.091 | undef | 0.083 |
|---|
| C6 |   | 4 | 440 | 913 | 904 | 0.128 | 0.055 | 0.169 | 0.161 | 0.054 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| SFRS17A |   | 3 | 557 | 913 | 908 | -0.036 | 0.169 | -0.225 | -0.176 | -0.078 |
|---|
| PECAM1 |   | 4 | 440 | 83 | 112 | 0.014 | -0.161 | 0.040 | -0.105 | -0.106 |
|---|
| PPIG |   | 8 | 222 | 648 | 631 | -0.116 | 0.024 | 0.221 | -0.065 | 0.101 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| PTPRZ1 |   | 9 | 196 | 570 | 554 | -0.114 | 0.183 | 0.233 | -0.316 | -0.095 |
|---|
| INSM1 |   | 23 | 66 | 570 | 545 | 0.135 | -0.184 | -0.188 | 0.237 | -0.032 |
|---|
| KCTD3 |   | 3 | 557 | 913 | 908 | 0.051 | 0.016 | 0.086 | 0.256 | 0.053 |
|---|
| MDFI |   | 17 | 95 | 236 | 226 | -0.075 | -0.001 | 0.177 | 0.169 | -0.112 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| FCGR2B |   | 2 | 743 | 913 | 914 | 0.253 | -0.024 | 0.051 | -0.284 | -0.002 |
|---|
| SOS1 |   | 5 | 360 | 478 | 476 | -0.049 | 0.002 | -0.007 | 0.160 | -0.042 |
|---|
| ZC4H2 |   | 17 | 95 | 570 | 546 | 0.065 | 0.061 | 0.304 | -0.072 | 0.059 |
|---|
| CHAF1B |   | 1 | 1195 | 913 | 953 | 0.015 | -0.193 | 0.171 | -0.052 | -0.041 |
|---|
| JMJD1C |   | 7 | 256 | 552 | 542 | -0.195 | 0.241 | 0.074 | undef | 0.129 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| SKP2 |   | 29 | 45 | 1 | 23 | 0.092 | -0.043 | 0.237 | undef | -0.009 |
|---|
| ITGA5 |   | 26 | 52 | 179 | 177 | 0.031 | 0.192 | 0.125 | 0.133 | 0.202 |
|---|
| NR2F1 |   | 25 | 59 | 83 | 92 | -0.252 | 0.113 | 0.133 | -0.116 | 0.207 |
|---|
| ARIH2 |   | 5 | 360 | 236 | 242 | -0.131 | 0.196 | -0.071 | -0.127 | -0.016 |
|---|
| CDK5R1 |   | 17 | 95 | 552 | 521 | -0.150 | 0.038 | -0.096 | 0.121 | 0.091 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| HIF1A |   | 13 | 124 | 236 | 230 | -0.098 | 0.224 | -0.118 | 0.135 | 0.356 |
|---|
| MEP1B |   | 2 | 743 | 913 | 914 | undef | 0.053 | 0.142 | 0.118 | -0.167 |
|---|
| TFDP1 |   | 6 | 301 | 806 | 783 | 0.123 | -0.178 | 0.129 | undef | -0.065 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| VWF |   | 5 | 360 | 525 | 518 | -0.091 | 0.072 | 0.152 | 0.127 | 0.100 |
|---|
| TIE1 |   | 18 | 86 | 366 | 357 | -0.108 | 0.005 | 0.141 | -0.108 | 0.252 |
|---|
| FOXA1 |   | 27 | 50 | 179 | 176 | -0.072 | 0.071 | -0.015 | 0.214 | 0.005 |
|---|
GO Enrichment output for subnetwork 6695 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| melatonin biosynthetic process | GO:0030187 |  | 3.538E-09 | 8.643E-06 |
|---|
| indolalkylamine biosynthetic process | GO:0046219 |  | 1.057E-08 | 1.291E-05 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 3.227E-07 | 2.628E-04 |
|---|
| indolalkylamine metabolic process | GO:0006586 |  | 4.896E-07 | 2.99E-04 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 1.377E-06 | 6.727E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.171E-06 | 1.698E-03 |
|---|
| prostate gland development | GO:0030850 |  | 4.781E-06 | 1.669E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 4.781E-06 | 1.46E-03 |
|---|
| ER overload response | GO:0006983 |  | 7.621E-06 | 2.069E-03 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 7.621E-06 | 1.862E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 7.621E-06 | 1.693E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| indolalkylamine biosynthetic process | GO:0046219 |  | 4.763E-09 | 1.146E-05 |
|---|
| indolalkylamine metabolic process | GO:0006586 |  | 3.062E-07 | 3.684E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 3.83E-07 | 3.072E-04 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 1.722E-06 | 1.036E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 5.966E-06 | 2.871E-03 |
|---|
| prostate gland development | GO:0030850 |  | 5.976E-06 | 2.396E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 5.976E-06 | 2.054E-03 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 9.314E-06 | 2.801E-03 |
|---|
| ER overload response | GO:0006983 |  | 9.522E-06 | 2.545E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 9.522E-06 | 2.291E-03 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 9.522E-06 | 2.083E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 3.65E-07 | 8.395E-04 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 3.282E-06 | 3.775E-03 |
|---|
| prostate gland development | GO:0030850 |  | 6.531E-06 | 5.007E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 6.531E-06 | 3.755E-03 |
|---|
| ER overload response | GO:0006983 |  | 6.531E-06 | 3.004E-03 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 6.531E-06 | 2.504E-03 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 8.069E-06 | 2.651E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.187E-05 | 3.412E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.81E-05 | 4.626E-03 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 1.81E-05 | 4.163E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 2.153E-05 | 4.502E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of glucose metabolic process | GO:0010907 |  | 4.03E-06 | 7.427E-03 |
|---|
| prostate gland development | GO:0030850 |  | 4.03E-06 | 3.714E-03 |
|---|
| positive regulation of catabolic process | GO:0009896 |  | 4.939E-06 | 3.034E-03 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 6.399E-06 | 2.948E-03 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 6.423E-06 | 2.368E-03 |
|---|
| cell junction assembly | GO:0034329 |  | 6.423E-06 | 1.973E-03 |
|---|
| ER overload response | GO:0006983 |  | 8.017E-06 | 2.111E-03 |
|---|
| regulation of cellular carbohydrate catabolic process | GO:0043471 |  | 8.017E-06 | 1.847E-03 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 9.607E-06 | 1.967E-03 |
|---|
| regulation of cell migration | GO:0030334 |  | 1.365E-05 | 2.515E-03 |
|---|
| positive regulation of cellular catabolic process | GO:0031331 |  | 1.396E-05 | 2.338E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 3.65E-07 | 8.395E-04 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 3.282E-06 | 3.775E-03 |
|---|
| prostate gland development | GO:0030850 |  | 6.531E-06 | 5.007E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 6.531E-06 | 3.755E-03 |
|---|
| ER overload response | GO:0006983 |  | 6.531E-06 | 3.004E-03 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 6.531E-06 | 2.504E-03 |
|---|
| positive regulation of fibroblast proliferation | GO:0048146 |  | 8.069E-06 | 2.651E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.187E-05 | 3.412E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.81E-05 | 4.626E-03 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 1.81E-05 | 4.163E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 2.153E-05 | 4.502E-03 |
|---|



