Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6681
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7519 | 3.640e-03 | 5.980e-04 | 7.756e-01 | 1.688e-06 |
|---|
| Loi | 0.2310 | 7.926e-02 | 1.110e-02 | 4.258e-01 | 3.745e-04 |
|---|
| Schmidt | 0.6743 | 0.000e+00 | 0.000e+00 | 4.402e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7909 | 0.000e+00 | 0.000e+00 | 3.457e-03 | 0.000e+00 |
|---|
| Wang | 0.2627 | 2.533e-03 | 4.248e-02 | 3.900e-01 | 4.196e-05 |
|---|
Expression data for subnetwork 6681 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
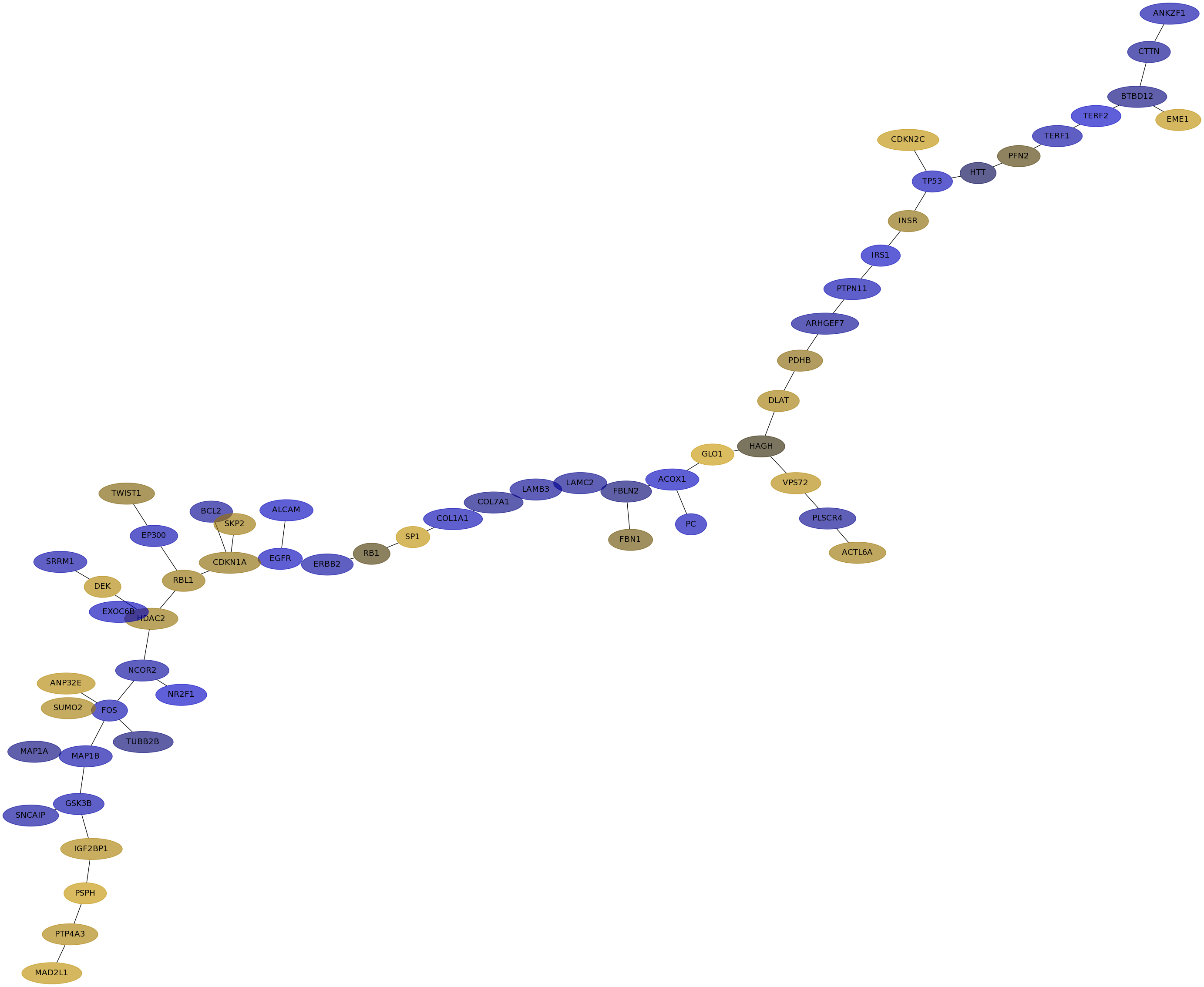
Score for each gene in subnetwork 6681 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| NCOR2 |   | 32 | 40 | 318 | 290 | -0.096 | 0.211 | -0.163 | 0.158 | 0.114 |
|---|
| PLSCR4 |   | 13 | 124 | 318 | 316 | -0.069 | 0.140 | 0.231 | 0.212 | 0.212 |
|---|
| FBLN2 |   | 5 | 360 | 83 | 110 | -0.036 | 0.122 | 0.207 | 0.037 | -0.007 |
|---|
| ARHGEF7 |   | 25 | 59 | 83 | 92 | -0.074 | -0.061 | 0.208 | 0.028 | 0.015 |
|---|
| SRRM1 |   | 10 | 167 | 296 | 283 | -0.120 | 0.139 | 0.244 | 0.294 | -0.109 |
|---|
| SUMO2 |   | 22 | 69 | 366 | 345 | 0.117 | 0.062 | 0.294 | 0.141 | -0.096 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| PTP4A3 |   | 9 | 196 | 412 | 411 | 0.137 | -0.196 | 0.085 | undef | -0.004 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PFN2 |   | 11 | 148 | 141 | 143 | 0.014 | -0.001 | 0.286 | -0.002 | -0.056 |
|---|
| LAMB3 |   | 1 | 1195 | 1247 | 1267 | -0.079 | 0.279 | -0.018 | -0.095 | -0.105 |
|---|
| CDKN2C |   | 16 | 104 | 236 | 227 | 0.216 | -0.106 | 0.202 | -0.024 | -0.081 |
|---|
| RB1 |   | 31 | 43 | 1 | 22 | 0.013 | -0.051 | 0.057 | 0.100 | -0.082 |
|---|
| BTBD12 |   | 1 | 1195 | 1247 | 1267 | -0.046 | 0.069 | undef | undef | undef |
|---|
| MAP1A |   | 12 | 140 | 1 | 36 | -0.045 | 0.143 | -0.054 | 0.162 | 0.147 |
|---|
| VPS72 |   | 6 | 301 | 318 | 330 | 0.164 | -0.182 | 0.095 | 0.151 | 0.080 |
|---|
| PTPN11 |   | 20 | 81 | 83 | 94 | -0.152 | 0.014 | 0.066 | 0.304 | 0.089 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| IGF2BP1 |   | 15 | 111 | 366 | 359 | 0.135 | -0.036 | undef | 0.190 | undef |
|---|
| CTTN |   | 2 | 743 | 1247 | 1238 | -0.063 | 0.232 | -0.064 | undef | 0.151 |
|---|
| TERF2 |   | 1 | 1195 | 1247 | 1267 | -0.252 | -0.034 | -0.033 | 0.029 | 0.262 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| EXOC6B |   | 6 | 301 | 318 | 330 | -0.183 | 0.205 | -0.104 | 0.030 | 0.037 |
|---|
| COL1A1 |   | 3 | 557 | 552 | 558 | -0.156 | 0.244 | -0.021 | -0.081 | 0.086 |
|---|
| SKP2 |   | 29 | 45 | 1 | 23 | 0.092 | -0.043 | 0.237 | undef | -0.009 |
|---|
| DEK |   | 1 | 1195 | 1247 | 1267 | 0.156 | -0.168 | 0.207 | undef | 0.093 |
|---|
| COL7A1 |   | 3 | 557 | 1222 | 1210 | -0.052 | 0.240 | 0.233 | -0.019 | 0.022 |
|---|
| GLO1 |   | 5 | 360 | 83 | 110 | 0.266 | -0.089 | 0.135 | 0.100 | 0.076 |
|---|
| HAGH |   | 2 | 743 | 1247 | 1238 | 0.007 | -0.155 | -0.128 | 0.202 | 0.033 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| ALCAM |   | 8 | 222 | 1 | 46 | -0.208 | 0.193 | 0.067 | 0.088 | -0.010 |
|---|
| PDHB |   | 13 | 124 | 525 | 503 | 0.056 | 0.017 | 0.197 | -0.205 | 0.193 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| NR2F1 |   | 25 | 59 | 83 | 92 | -0.252 | 0.113 | 0.133 | -0.116 | 0.207 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| IRS1 |   | 9 | 196 | 842 | 805 | -0.219 | 0.065 | 0.165 | 0.102 | 0.064 |
|---|
| ANKZF1 |   | 1 | 1195 | 1247 | 1267 | -0.119 | 0.182 | 0.045 | 0.109 | 0.060 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| PC |   | 1 | 1195 | 1247 | 1267 | -0.171 | -0.017 | -0.024 | 0.236 | -0.060 |
|---|
| FBN1 |   | 1 | 1195 | 1247 | 1267 | 0.030 | 0.085 | 0.060 | undef | 0.157 |
|---|
| HTT |   | 14 | 117 | 141 | 140 | -0.016 | -0.015 | 0.075 | 0.092 | -0.093 |
|---|
| RBL1 |   | 21 | 73 | 1 | 27 | 0.078 | undef | undef | 0.051 | undef |
|---|
| ACOX1 |   | 9 | 196 | 1 | 44 | -0.209 | 0.122 | -0.104 | 0.252 | -0.094 |
|---|
| MAD2L1 |   | 8 | 222 | 236 | 236 | 0.208 | -0.130 | 0.116 | 0.032 | 0.063 |
|---|
| TUBB2B |   | 13 | 124 | 141 | 141 | -0.036 | 0.304 | 0.045 | 0.187 | 0.139 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| ANP32E |   | 2 | 743 | 1247 | 1238 | 0.166 | -0.139 | 0.191 | undef | 0.118 |
|---|
| EME1 |   | 1 | 1195 | 1247 | 1267 | 0.209 | 0.119 | undef | undef | undef |
|---|
| SNCAIP |   | 10 | 167 | 366 | 362 | -0.091 | 0.013 | 0.092 | 0.136 | 0.059 |
|---|
| TERF1 |   | 1 | 1195 | 1247 | 1267 | -0.107 | 0.006 | 0.111 | 0.222 | 0.126 |
|---|
| ACTL6A |   | 2 | 743 | 1247 | 1238 | 0.095 | -0.069 | 0.193 | undef | 0.040 |
|---|
| LAMC2 |   | 4 | 440 | 179 | 209 | -0.063 | 0.307 | -0.060 | -0.148 | 0.055 |
|---|
| PSPH |   | 20 | 81 | 366 | 356 | 0.240 | -0.019 | -0.102 | 0.358 | 0.025 |
|---|
| HDAC2 |   | 13 | 124 | 318 | 316 | 0.079 | -0.094 | 0.148 | 0.089 | 0.037 |
|---|
| DLAT |   | 4 | 440 | 1141 | 1127 | 0.111 | 0.029 | 0.072 | undef | -0.014 |
|---|
GO Enrichment output for subnetwork 6681 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 2.277E-11 | 5.562E-08 |
|---|
| organ growth | GO:0035265 |  | 2.277E-11 | 2.781E-08 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.989E-10 | 6.505E-07 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 9.469E-09 | 5.783E-06 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 1.822E-08 | 8.903E-06 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.888E-08 | 7.687E-06 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 1.888E-08 | 6.589E-06 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 3.387E-08 | 1.034E-05 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.204E-07 | 3.268E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.728E-07 | 4.222E-05 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 3.208E-07 | 7.126E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 2.781E-11 | 6.691E-08 |
|---|
| organ growth | GO:0035265 |  | 2.781E-11 | 3.346E-08 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.045E-09 | 8.384E-07 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 1.113E-08 | 6.696E-06 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 1.942E-08 | 9.345E-06 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 2.219E-08 | 8.898E-06 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 3.981E-08 | 1.368E-05 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.566E-07 | 4.71E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.814E-07 | 4.849E-05 |
|---|
| interphase | GO:0051325 |  | 2.247E-07 | 5.406E-05 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 2.875E-07 | 6.287E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 2.545E-11 | 5.854E-08 |
|---|
| organ growth | GO:0035265 |  | 8.87E-11 | 1.02E-07 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.071E-09 | 2.355E-06 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 4.074E-09 | 2.343E-06 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 2.828E-08 | 1.301E-05 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 5.397E-08 | 2.069E-05 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 5.633E-08 | 1.851E-05 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 3.909E-07 | 1.124E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.237E-07 | 1.083E-04 |
|---|
| interphase | GO:0051325 |  | 4.962E-07 | 1.141E-04 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 6.474E-07 | 1.354E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.55E-08 | 1.576E-04 |
|---|
| negative regulation of homeostatic process | GO:0032845 |  | 1.807E-07 | 1.665E-04 |
|---|
| regulation of chromosome organization | GO:0033044 |  | 4.225E-07 | 2.595E-04 |
|---|
| negative regulation of DNA replication | GO:0008156 |  | 6.078E-07 | 2.8E-04 |
|---|
| regulation of DNA replication | GO:0006275 |  | 6.85E-07 | 2.525E-04 |
|---|
| negative regulation of cell growth | GO:0030308 |  | 8.085E-07 | 2.483E-04 |
|---|
| negative regulation of cell size | GO:0045792 |  | 9.49E-07 | 2.498E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.489E-06 | 3.43E-04 |
|---|
| T cell lineage commitment | GO:0002360 |  | 1.671E-06 | 3.422E-04 |
|---|
| negative regulation of growth | GO:0045926 |  | 1.714E-06 | 3.16E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 2.246E-06 | 3.763E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 2.545E-11 | 5.854E-08 |
|---|
| organ growth | GO:0035265 |  | 8.87E-11 | 1.02E-07 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.071E-09 | 2.355E-06 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 4.074E-09 | 2.343E-06 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 2.828E-08 | 1.301E-05 |
|---|
| cell cycle checkpoint | GO:0000075 |  | 5.397E-08 | 2.069E-05 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 5.633E-08 | 1.851E-05 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 3.909E-07 | 1.124E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.237E-07 | 1.083E-04 |
|---|
| interphase | GO:0051325 |  | 4.962E-07 | 1.141E-04 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 6.474E-07 | 1.354E-04 |
|---|



