Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6660
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7523 | 3.616e-03 | 5.930e-04 | 7.747e-01 | 1.661e-06 |
|---|
| Loi | 0.2309 | 7.940e-02 | 1.113e-02 | 4.261e-01 | 3.767e-04 |
|---|
| Schmidt | 0.6741 | 0.000e+00 | 0.000e+00 | 4.420e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.476e-03 | 0.000e+00 |
|---|
| Wang | 0.2626 | 2.547e-03 | 4.260e-02 | 3.906e-01 | 4.239e-05 |
|---|
Expression data for subnetwork 6660 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
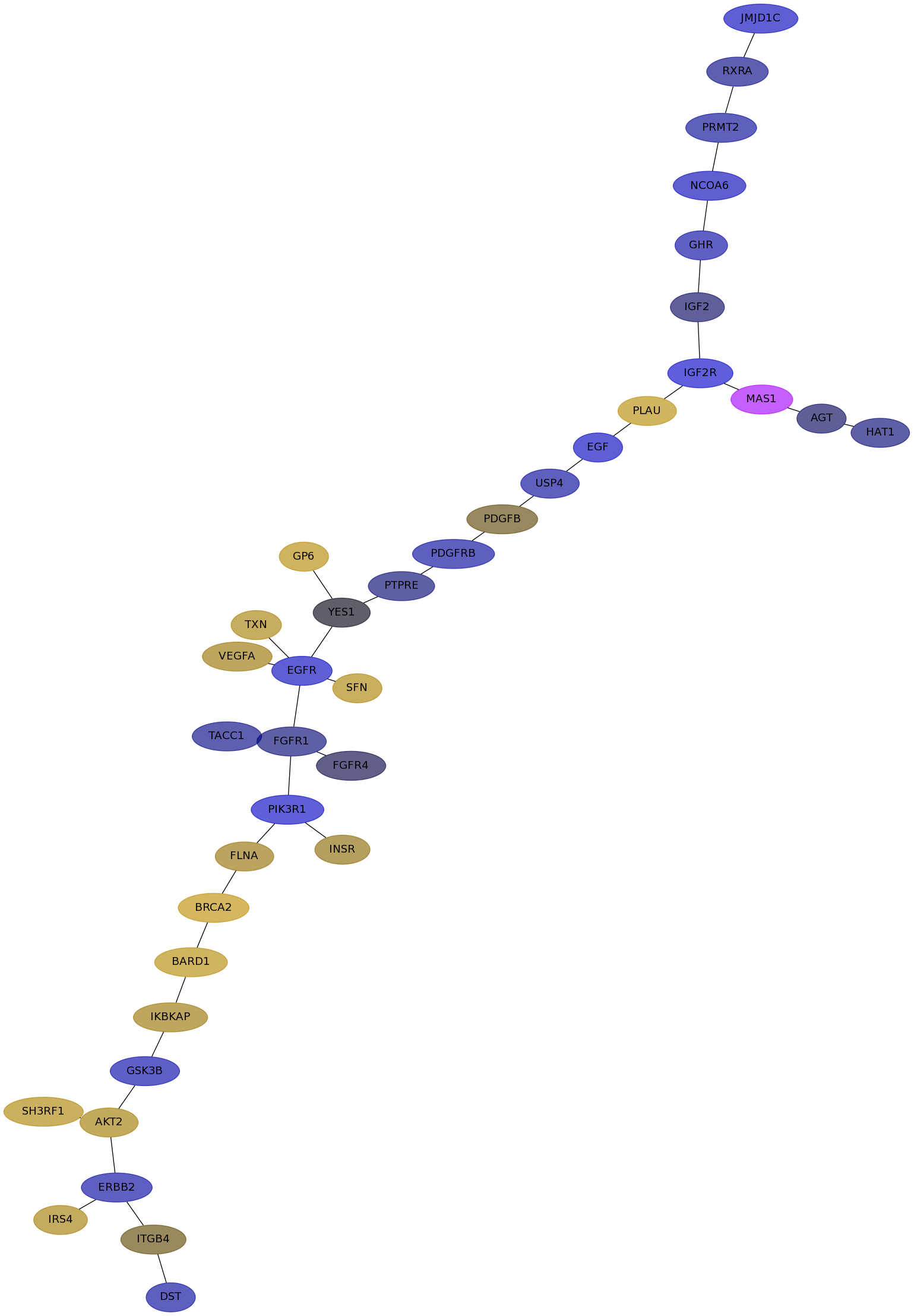
Score for each gene in subnetwork 6660 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| USP4 |   | 7 | 256 | 1 | 47 | -0.093 | 0.147 | 0.097 | -0.392 | 0.054 |
|---|
| TXN |   | 2 | 743 | 1486 | 1473 | 0.124 | 0.012 | 0.014 | 0.006 | -0.006 |
|---|
| MAS1 |   | 1 | 1195 | 1486 | 1511 | undef | -0.072 | -0.041 | 0.173 | 0.046 |
|---|
| EGF |   | 12 | 140 | 638 | 620 | -0.212 | 0.182 | -0.276 | -0.060 | 0.005 |
|---|
| RXRA |   | 7 | 256 | 457 | 451 | -0.055 | 0.134 | -0.027 | 0.022 | -0.096 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| IKBKAP |   | 10 | 167 | 412 | 408 | 0.089 | 0.184 | 0.142 | 0.105 | -0.017 |
|---|
| PDGFB |   | 7 | 256 | 1165 | 1135 | 0.020 | 0.046 | 0.085 | -0.049 | 0.338 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| DST |   | 22 | 69 | 1 | 25 | -0.094 | 0.343 | 0.065 | -0.062 | 0.194 |
|---|
| PLAU |   | 12 | 140 | 179 | 192 | 0.187 | 0.153 | 0.141 | 0.072 | 0.192 |
|---|
| NCOA6 |   | 3 | 557 | 957 | 949 | -0.179 | -0.123 | -0.167 | 0.103 | 0.133 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| HAT1 |   | 1 | 1195 | 1486 | 1511 | -0.035 | -0.052 | 0.073 | 0.049 | -0.040 |
|---|
| PTPRE |   | 1 | 1195 | 1486 | 1511 | -0.032 | 0.044 | 0.059 | 0.058 | 0.046 |
|---|
| PDGFRB |   | 12 | 140 | 141 | 142 | -0.094 | 0.224 | 0.127 | 0.153 | 0.189 |
|---|
| FGFR4 |   | 21 | 73 | 179 | 183 | -0.011 | 0.188 | -0.089 | -0.018 | -0.049 |
|---|
| IGF2R |   | 2 | 743 | 638 | 650 | -0.264 | -0.027 | -0.031 | 0.205 | 0.005 |
|---|
| TACC1 |   | 5 | 360 | 1077 | 1057 | -0.051 | 0.020 | -0.022 | 0.187 | 0.126 |
|---|
| AKT2 |   | 28 | 47 | 412 | 394 | 0.110 | -0.118 | -0.003 | 0.088 | -0.092 |
|---|
| JMJD1C |   | 7 | 256 | 552 | 542 | -0.195 | 0.241 | 0.074 | undef | 0.129 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| YES1 |   | 1 | 1195 | 1486 | 1511 | -0.002 | -0.202 | 0.075 | 0.051 | 0.185 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| PRMT2 |   | 5 | 360 | 957 | 938 | -0.081 | 0.046 | 0.089 | 0.235 | -0.128 |
|---|
| IRS4 |   | 32 | 40 | 236 | 224 | 0.115 | -0.111 | -0.088 | 0.096 | 0.142 |
|---|
| FGFR1 |   | 26 | 52 | 638 | 615 | -0.036 | 0.200 | 0.087 | undef | -0.015 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| AGT |   | 4 | 440 | 1203 | 1184 | -0.020 | -0.101 | -0.043 | 0.255 | 0.064 |
|---|
| SH3RF1 |   | 6 | 301 | 1222 | 1194 | 0.144 | 0.238 | undef | undef | undef |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| GP6 |   | 1 | 1195 | 1486 | 1511 | 0.170 | 0.147 | -0.098 | undef | -0.005 |
|---|
| ITGB4 |   | 35 | 34 | 1 | 20 | 0.021 | 0.313 | -0.134 | 0.080 | -0.114 |
|---|
| IGF2 |   | 14 | 117 | 570 | 553 | -0.022 | 0.172 | 0.145 | 0.087 | 0.116 |
|---|
| SFN |   | 25 | 59 | 478 | 454 | 0.141 | 0.270 | 0.023 | -0.102 | 0.101 |
|---|
GO Enrichment output for subnetwork 6660 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 4.103E-11 | 1.002E-07 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 9.137E-10 | 1.116E-06 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 2.076E-09 | 1.69E-06 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 3.182E-09 | 1.944E-06 |
|---|
| positive regulation of endothelial cell migration | GO:0010595 |  | 7.726E-09 | 3.775E-06 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 2.619E-08 | 1.066E-05 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 2.567E-07 | 8.96E-05 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 3.459E-07 | 1.056E-04 |
|---|
| regulation of endothelial cell migration | GO:0010594 |  | 4.16E-07 | 1.129E-04 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.205E-06 | 2.944E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.846E-06 | 4.099E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 2.672E-11 | 6.43E-08 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 8.901E-10 | 1.071E-06 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 2.052E-09 | 1.646E-06 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 2.849E-09 | 1.714E-06 |
|---|
| positive regulation of endothelial cell migration | GO:0010595 |  | 8.254E-09 | 3.972E-06 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 2.891E-08 | 1.159E-05 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 2.742E-07 | 9.424E-05 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 3.643E-07 | 1.096E-04 |
|---|
| regulation of endothelial cell migration | GO:0010594 |  | 4.442E-07 | 1.188E-04 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.269E-06 | 3.053E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.683E-06 | 3.681E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 2.05E-11 | 4.715E-08 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 2.644E-09 | 3.041E-06 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 5.203E-09 | 3.989E-06 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 1.211E-08 | 6.962E-06 |
|---|
| positive regulation of endothelial cell migration | GO:0010595 |  | 2.532E-08 | 1.165E-05 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 7.544E-08 | 2.892E-05 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 9.988E-08 | 3.282E-05 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 8.49E-07 | 2.441E-04 |
|---|
| regulation of endothelial cell migration | GO:0010594 |  | 1.351E-06 | 3.452E-04 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.693E-06 | 3.893E-04 |
|---|
| protein heterooligomerization | GO:0051291 |  | 2.952E-06 | 6.173E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell migration | GO:0030335 |  | 6.336E-11 | 1.168E-07 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.158E-10 | 1.067E-07 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 9.09E-10 | 5.584E-07 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 2.163E-09 | 9.964E-07 |
|---|
| regulation of cell migration | GO:0030334 |  | 9.188E-09 | 3.387E-06 |
|---|
| positive regulation of endothelial cell migration | GO:0010595 |  | 1.519E-08 | 4.667E-06 |
|---|
| regulation of locomotion | GO:0040012 |  | 1.96E-08 | 5.16E-06 |
|---|
| regulation of cell motion | GO:0051270 |  | 1.96E-08 | 4.515E-06 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 2.727E-08 | 5.585E-06 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 3.185E-08 | 5.869E-06 |
|---|
| regulation of protein amino acid phosphorylation | GO:0001932 |  | 1.223E-07 | 2.05E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 2.05E-11 | 4.715E-08 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 2.644E-09 | 3.041E-06 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 5.203E-09 | 3.989E-06 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 1.211E-08 | 6.962E-06 |
|---|
| positive regulation of endothelial cell migration | GO:0010595 |  | 2.532E-08 | 1.165E-05 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 7.544E-08 | 2.892E-05 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 9.988E-08 | 3.282E-05 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 8.49E-07 | 2.441E-04 |
|---|
| regulation of endothelial cell migration | GO:0010594 |  | 1.351E-06 | 3.452E-04 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.693E-06 | 3.893E-04 |
|---|
| protein heterooligomerization | GO:0051291 |  | 2.952E-06 | 6.173E-04 |
|---|



