Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6658
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7524 | 3.611e-03 | 5.920e-04 | 7.745e-01 | 1.656e-06 |
|---|
| Loi | 0.2308 | 7.957e-02 | 1.118e-02 | 4.265e-01 | 3.793e-04 |
|---|
| Schmidt | 0.6741 | 0.000e+00 | 0.000e+00 | 4.415e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.478e-03 | 0.000e+00 |
|---|
| Wang | 0.2625 | 2.553e-03 | 4.266e-02 | 3.909e-01 | 4.258e-05 |
|---|
Expression data for subnetwork 6658 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
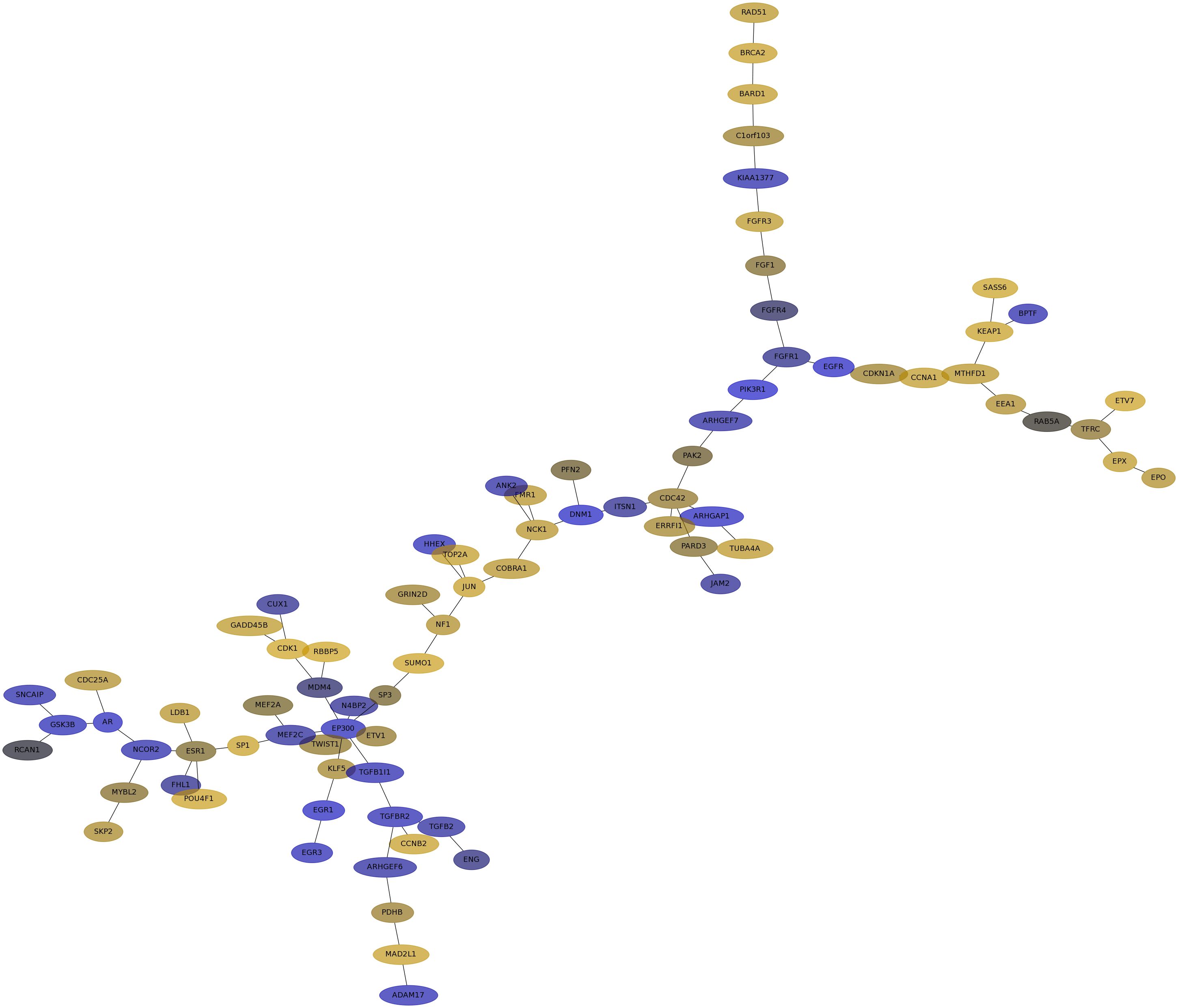
Score for each gene in subnetwork 6658 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| NF1 |   | 9 | 196 | 552 | 541 | 0.110 | -0.052 | -0.032 | 0.090 | -0.094 |
|---|
| BPTF |   | 4 | 440 | 318 | 337 | -0.098 | 0.159 | 0.132 | undef | 0.030 |
|---|
| MDM4 |   | 9 | 196 | 141 | 147 | -0.016 | 0.004 | 0.164 | 0.107 | 0.105 |
|---|
| EPX |   | 3 | 557 | 478 | 487 | 0.172 | -0.224 | -0.005 | 0.173 | -0.064 |
|---|
| EGR3 |   | 1 | 1195 | 1604 | 1620 | -0.124 | -0.026 | 0.130 | undef | -0.113 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| EEA1 |   | 1 | 1195 | 1604 | 1620 | 0.102 | -0.028 | -0.043 | 0.022 | 0.004 |
|---|
| ENG |   | 2 | 743 | 842 | 854 | -0.027 | 0.062 | 0.130 | 0.174 | 0.010 |
|---|
| CDC42 |   | 26 | 52 | 179 | 177 | 0.047 | 0.242 | -0.027 | 0.040 | -0.042 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| ETV1 |   | 10 | 167 | 1 | 40 | 0.046 | 0.054 | -0.065 | 0.014 | 0.141 |
|---|
| ITSN1 |   | 1 | 1195 | 1604 | 1620 | -0.044 | 0.026 | 0.113 | 0.064 | 0.124 |
|---|
| TUBA4A |   | 7 | 256 | 1 | 47 | 0.157 | 0.136 | -0.010 | undef | 0.156 |
|---|
| COBRA1 |   | 2 | 743 | 1604 | 1588 | 0.137 | 0.335 | 0.159 | 0.134 | -0.016 |
|---|
| SP3 |   | 1 | 1195 | 1604 | 1620 | 0.020 | 0.088 | 0.028 | undef | 0.112 |
|---|
| NCK1 |   | 3 | 557 | 318 | 341 | 0.135 | -0.100 | -0.059 | -0.108 | 0.151 |
|---|
| ANK2 |   | 3 | 557 | 1604 | 1577 | -0.073 | -0.023 | 0.095 | -0.019 | 0.095 |
|---|
| CUX1 |   | 6 | 301 | 1203 | 1169 | -0.042 | 0.256 | -0.121 | 0.105 | 0.040 |
|---|
| POU4F1 |   | 1 | 1195 | 1604 | 1620 | 0.247 | 0.031 | 0.177 | -0.137 | -0.027 |
|---|
| RAB5A |   | 2 | 743 | 1351 | 1344 | 0.002 | 0.087 | 0.154 | -0.093 | 0.045 |
|---|
| PDHB |   | 13 | 124 | 525 | 503 | 0.056 | 0.017 | 0.197 | -0.205 | 0.193 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| FGFR1 |   | 26 | 52 | 638 | 615 | -0.036 | 0.200 | 0.087 | undef | -0.015 |
|---|
| FGF1 |   | 4 | 440 | 1604 | 1572 | 0.027 | undef | undef | 0.090 | undef |
|---|
| HHEX |   | 4 | 440 | 727 | 723 | -0.130 | -0.178 | 0.177 | -0.180 | 0.005 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| ARHGEF6 |   | 10 | 167 | 525 | 507 | -0.068 | -0.120 | 0.166 | undef | -0.115 |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| GADD45B |   | 8 | 222 | 318 | 325 | 0.158 | 0.122 | 0.067 | 0.060 | 0.212 |
|---|
| MAD2L1 |   | 8 | 222 | 236 | 236 | 0.208 | -0.130 | 0.116 | 0.032 | 0.063 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| SASS6 |   | 1 | 1195 | 1604 | 1620 | 0.238 | -0.202 | undef | undef | undef |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| PARD3 |   | 6 | 301 | 366 | 377 | 0.030 | -0.006 | -0.052 | 0.040 | -0.090 |
|---|
| MEF2C |   | 1 | 1195 | 1604 | 1620 | -0.062 | -0.097 | 0.112 | -0.189 | 0.132 |
|---|
| NCOR2 |   | 32 | 40 | 318 | 290 | -0.096 | 0.211 | -0.163 | 0.158 | 0.114 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| ARHGEF7 |   | 25 | 59 | 83 | 92 | -0.074 | -0.061 | 0.208 | 0.028 | 0.015 |
|---|
| MEF2A |   | 4 | 440 | 614 | 618 | 0.022 | 0.036 | 0.265 | -0.094 | 0.124 |
|---|
| RCAN1 |   | 2 | 743 | 366 | 395 | -0.002 | 0.111 | 0.005 | -0.019 | -0.045 |
|---|
| KIAA1377 |   | 18 | 86 | 141 | 136 | -0.095 | 0.116 | undef | 0.198 | undef |
|---|
| TGFB2 |   | 7 | 256 | 525 | 515 | -0.063 | 0.244 | 0.226 | 0.079 | 0.015 |
|---|
| PFN2 |   | 11 | 148 | 141 | 143 | 0.014 | -0.001 | 0.286 | -0.002 | -0.056 |
|---|
| TGFBR2 |   | 7 | 256 | 525 | 515 | -0.115 | -0.028 | 0.136 | 0.105 | -0.190 |
|---|
| RBBP5 |   | 3 | 557 | 478 | 487 | 0.264 | 0.053 | 0.086 | undef | 0.033 |
|---|
| TFRC |   | 5 | 360 | 957 | 938 | 0.038 | -0.066 | 0.076 | -0.108 | -0.014 |
|---|
| SUMO1 |   | 5 | 360 | 1034 | 1004 | 0.245 | -0.007 | 0.144 | 0.083 | 0.118 |
|---|
| TOP2A |   | 20 | 81 | 1 | 28 | 0.193 | -0.168 | 0.054 | -0.064 | 0.071 |
|---|
| JUN |   | 26 | 52 | 179 | 177 | 0.200 | 0.051 | 0.220 | undef | 0.200 |
|---|
| FGFR3 |   | 9 | 196 | 179 | 195 | 0.160 | 0.071 | 0.018 | -0.050 | -0.111 |
|---|
| PAK2 |   | 26 | 52 | 179 | 177 | 0.014 | 0.161 | 0.081 | -0.044 | 0.062 |
|---|
| ARHGAP1 |   | 6 | 301 | 179 | 205 | -0.167 | 0.297 | 0.001 | -0.032 | 0.251 |
|---|
| ERRFI1 |   | 10 | 167 | 412 | 408 | 0.076 | 0.215 | undef | -0.043 | undef |
|---|
| ETV7 |   | 4 | 440 | 957 | 942 | 0.245 | -0.201 | -0.043 | -0.062 | -0.174 |
|---|
| C1orf103 |   | 4 | 440 | 412 | 419 | 0.057 | 0.029 | -0.173 | 0.127 | -0.159 |
|---|
| JAM2 |   | 2 | 743 | 1604 | 1588 | -0.047 | 0.075 | 0.241 | -0.215 | 0.055 |
|---|
| FMR1 |   | 2 | 743 | 1604 | 1588 | 0.137 | -0.104 | 0.352 | -0.014 | 0.113 |
|---|
| N4BP2 |   | 1 | 1195 | 1604 | 1620 | -0.046 | 0.054 | undef | undef | undef |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| MTHFD1 |   | 2 | 743 | 1604 | 1588 | 0.139 | -0.266 | 0.078 | 0.066 | 0.124 |
|---|
| FGFR4 |   | 21 | 73 | 179 | 183 | -0.011 | 0.188 | -0.089 | -0.018 | -0.049 |
|---|
| DNM1 |   | 4 | 440 | 236 | 262 | -0.203 | 0.095 | 0.163 | 0.001 | 0.082 |
|---|
| CDC25A |   | 8 | 222 | 570 | 555 | 0.113 | -0.170 | 0.151 | -0.301 | -0.003 |
|---|
| ADAM17 |   | 1 | 1195 | 1604 | 1620 | -0.113 | 0.058 | -0.073 | -0.081 | -0.106 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| LDB1 |   | 2 | 743 | 1323 | 1313 | 0.138 | -0.079 | -0.010 | -0.051 | -0.100 |
|---|
| MYBL2 |   | 4 | 440 | 1604 | 1572 | 0.031 | -0.220 | 0.034 | -0.034 | 0.006 |
|---|
| SKP2 |   | 29 | 45 | 1 | 23 | 0.092 | -0.043 | 0.237 | undef | -0.009 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| KEAP1 |   | 2 | 743 | 1604 | 1588 | 0.218 | 0.011 | -0.155 | -0.205 | -0.031 |
|---|
| RAD51 |   | 13 | 124 | 727 | 699 | 0.146 | -0.098 | 0.135 | -0.070 | 0.061 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| GRIN2D |   | 13 | 124 | 552 | 522 | 0.058 | 0.155 | 0.000 | undef | undef |
|---|
| EPO |   | 4 | 440 | 1492 | 1461 | 0.109 | 0.086 | -0.083 | -0.048 | 0.155 |
|---|
| FHL1 |   | 14 | 117 | 614 | 591 | -0.041 | 0.104 | 0.301 | undef | 0.266 |
|---|
| CCNB2 |   | 4 | 440 | 842 | 831 | 0.206 | -0.119 | 0.004 | -0.006 | 0.134 |
|---|
| ESR1 |   | 4 | 440 | 179 | 209 | 0.026 | -0.002 | -0.053 | -0.149 | -0.062 |
|---|
| SNCAIP |   | 10 | 167 | 366 | 362 | -0.091 | 0.013 | 0.092 | 0.136 | 0.059 |
|---|
GO Enrichment output for subnetwork 6658 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 1.154E-10 | 2.818E-07 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 2.313E-10 | 2.826E-07 |
|---|
| lung development | GO:0030324 |  | 1.334E-09 | 1.086E-06 |
|---|
| respiratory tube development | GO:0030323 |  | 1.967E-09 | 1.202E-06 |
|---|
| respiratory system development | GO:0060541 |  | 2.524E-09 | 1.233E-06 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 2.677E-08 | 1.09E-05 |
|---|
| cell growth | GO:0016049 |  | 3.139E-08 | 1.096E-05 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 4.51E-08 | 1.377E-05 |
|---|
| inner ear development | GO:0048839 |  | 8.673E-08 | 2.354E-05 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 9.177E-08 | 2.242E-05 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 1.264E-07 | 2.806E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 8.184E-11 | 1.969E-07 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 1.778E-10 | 2.139E-07 |
|---|
| lung development | GO:0030324 |  | 3.696E-08 | 2.964E-05 |
|---|
| cell growth | GO:0016049 |  | 4.091E-08 | 2.461E-05 |
|---|
| respiratory tube development | GO:0030323 |  | 5.248E-08 | 2.525E-05 |
|---|
| respiratory system development | GO:0060541 |  | 6.57E-08 | 2.634E-05 |
|---|
| inner ear development | GO:0048839 |  | 8.725E-08 | 2.999E-05 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 8.893E-08 | 2.675E-05 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 1.252E-07 | 3.348E-05 |
|---|
| negative regulation of epithelial cell proliferation | GO:0050680 |  | 2.325E-07 | 5.593E-05 |
|---|
| regulation of protein complex assembly | GO:0043254 |  | 3.556E-07 | 7.777E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell growth | GO:0016049 |  | 7.928E-08 | 1.824E-04 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 9.338E-08 | 1.074E-04 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 3.096E-07 | 2.374E-04 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 5.979E-07 | 3.438E-04 |
|---|
| negative regulation of epithelial cell proliferation | GO:0050680 |  | 8.051E-07 | 3.704E-04 |
|---|
| regulation of protein complex assembly | GO:0043254 |  | 8.088E-07 | 3.101E-04 |
|---|
| myeloid leukocyte differentiation | GO:0002573 |  | 1.387E-06 | 4.558E-04 |
|---|
| protein sumoylation | GO:0016925 |  | 1.652E-06 | 4.749E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 1.807E-06 | 4.618E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.831E-06 | 6.511E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.301E-06 | 8.993E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.374E-06 | 2.532E-03 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 2.098E-06 | 1.933E-03 |
|---|
| tube development | GO:0035295 |  | 2.106E-06 | 1.294E-03 |
|---|
| heart development | GO:0007507 |  | 3.418E-06 | 1.575E-03 |
|---|
| lung development | GO:0030324 |  | 7.357E-06 | 2.712E-03 |
|---|
| artery development | GO:0060840 |  | 8.017E-06 | 2.463E-03 |
|---|
| positive regulation of cellular component organization | GO:0051130 |  | 8.148E-06 | 2.145E-03 |
|---|
| respiratory tube development | GO:0030323 |  | 9.607E-06 | 2.213E-03 |
|---|
| respiratory system development | GO:0060541 |  | 9.607E-06 | 1.967E-03 |
|---|
| induction of an organ | GO:0001759 |  | 1.396E-05 | 2.572E-03 |
|---|
| developmental induction | GO:0031128 |  | 2.221E-05 | 3.721E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell growth | GO:0016049 |  | 7.928E-08 | 1.824E-04 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 9.338E-08 | 1.074E-04 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 3.096E-07 | 2.374E-04 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 5.979E-07 | 3.438E-04 |
|---|
| negative regulation of epithelial cell proliferation | GO:0050680 |  | 8.051E-07 | 3.704E-04 |
|---|
| regulation of protein complex assembly | GO:0043254 |  | 8.088E-07 | 3.101E-04 |
|---|
| myeloid leukocyte differentiation | GO:0002573 |  | 1.387E-06 | 4.558E-04 |
|---|
| protein sumoylation | GO:0016925 |  | 1.652E-06 | 4.749E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 1.807E-06 | 4.618E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.831E-06 | 6.511E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.301E-06 | 8.993E-04 |
|---|



