Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6634
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7530 | 3.570e-03 | 5.840e-04 | 7.729e-01 | 1.611e-06 |
|---|
| Loi | 0.2304 | 8.006e-02 | 1.130e-02 | 4.277e-01 | 3.871e-04 |
|---|
| Schmidt | 0.6742 | 0.000e+00 | 0.000e+00 | 4.405e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.464e-03 | 0.000e+00 |
|---|
| Wang | 0.2624 | 2.564e-03 | 4.277e-02 | 3.914e-01 | 4.292e-05 |
|---|
Expression data for subnetwork 6634 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
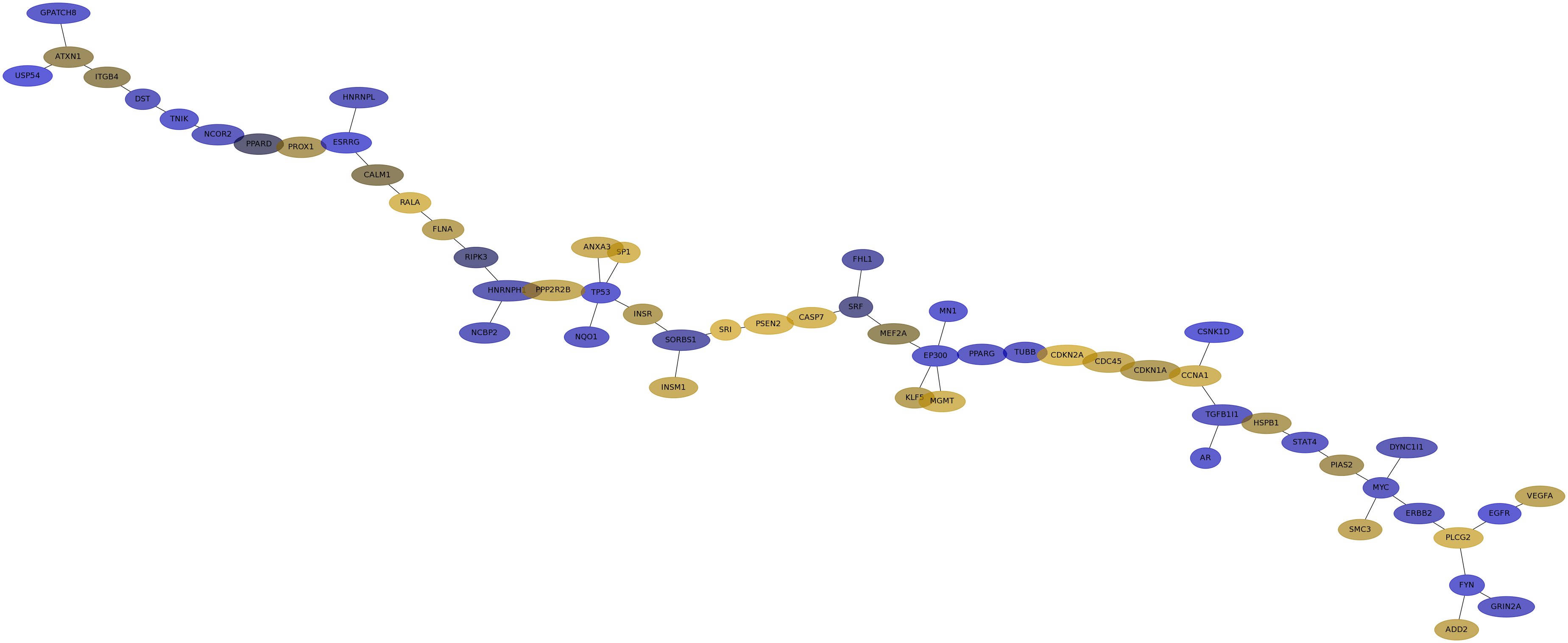
Score for each gene in subnetwork 6634 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| ANXA3 |   | 3 | 557 | 141 | 170 | 0.152 | 0.132 | 0.194 | 0.090 | 0.030 |
|---|
| TNIK |   | 3 | 557 | 614 | 623 | -0.156 | 0.084 | undef | 0.072 | -0.090 |
|---|
| NCOR2 |   | 32 | 40 | 318 | 290 | -0.096 | 0.211 | -0.163 | 0.158 | 0.114 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| NQO1 |   | 21 | 73 | 366 | 354 | -0.115 | 0.069 | -0.189 | 0.321 | 0.118 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ADD2 |   | 8 | 222 | 614 | 599 | 0.113 | -0.022 | -0.061 | 0.270 | 0.180 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| MEF2A |   | 4 | 440 | 614 | 618 | 0.022 | 0.036 | 0.265 | -0.094 | 0.124 |
|---|
| HNRNPH1 |   | 9 | 196 | 614 | 597 | -0.059 | 0.248 | 0.125 | 0.144 | -0.133 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| DYNC1I1 |   | 5 | 360 | 614 | 612 | -0.069 | 0.013 | -0.086 | -0.045 | 0.088 |
|---|
| DST |   | 22 | 69 | 1 | 25 | -0.094 | 0.343 | 0.065 | -0.062 | 0.194 |
|---|
| SMC3 |   | 5 | 360 | 614 | 612 | 0.105 | -0.096 | 0.238 | 0.137 | 0.047 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| GPATCH8 |   | 8 | 222 | 614 | 599 | -0.143 | 0.014 | 0.019 | -0.070 | 0.061 |
|---|
| PLCG2 |   | 1 | 1195 | 614 | 660 | 0.202 | -0.199 | -0.011 | -0.339 | -0.112 |
|---|
| PSEN2 |   | 14 | 117 | 614 | 591 | 0.239 | -0.068 | -0.157 | 0.295 | 0.045 |
|---|
| SRI |   | 7 | 256 | 614 | 601 | 0.270 | -0.040 | 0.181 | 0.185 | 0.057 |
|---|
| USP54 |   | 1 | 1195 | 614 | 660 | -0.245 | 0.275 | undef | undef | undef |
|---|
| INSM1 |   | 23 | 66 | 570 | 545 | 0.135 | -0.184 | -0.188 | 0.237 | -0.032 |
|---|
| HNRNPL |   | 2 | 743 | 614 | 632 | -0.086 | -0.018 | 0.046 | 0.178 | 0.118 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| GRIN2A |   | 12 | 140 | 179 | 192 | -0.113 | -0.068 | 0.162 | 0.125 | undef |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| MN1 |   | 3 | 557 | 366 | 389 | -0.166 | -0.047 | 0.150 | 0.010 | 0.011 |
|---|
| PPARG |   | 7 | 256 | 179 | 201 | -0.119 | 0.011 | 0.105 | 0.059 | 0.039 |
|---|
| ESRRG |   | 6 | 301 | 614 | 602 | -0.205 | 0.197 | -0.007 | 0.155 | -0.085 |
|---|
| STAT4 |   | 1 | 1195 | 614 | 660 | -0.118 | -0.179 | -0.025 | -0.184 | -0.047 |
|---|
| RIPK3 |   | 19 | 85 | 179 | 186 | -0.014 | 0.040 | undef | 0.173 | undef |
|---|
| CALM1 |   | 6 | 301 | 614 | 602 | 0.014 | 0.021 | 0.192 | 0.221 | 0.247 |
|---|
| PROX1 |   | 4 | 440 | 614 | 618 | 0.050 | -0.030 | -0.128 | 0.161 | 0.112 |
|---|
| ATXN1 |   | 17 | 95 | 1 | 29 | 0.026 | 0.192 | 0.175 | 0.125 | 0.163 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| MGMT |   | 5 | 360 | 366 | 379 | 0.184 | -0.022 | 0.201 | undef | 0.196 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| CDC45 |   | 5 | 360 | 318 | 334 | 0.134 | -0.171 | 0.185 | -0.103 | -0.085 |
|---|
| RALA |   | 10 | 167 | 318 | 323 | 0.224 | 0.207 | -0.047 | 0.169 | 0.146 |
|---|
| PPARD |   | 2 | 743 | 614 | 632 | -0.006 | 0.091 | -0.002 | 0.130 | 0.194 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| CASP7 |   | 6 | 301 | 614 | 602 | 0.216 | 0.095 | 0.255 | undef | 0.035 |
|---|
| FYN |   | 10 | 167 | 1 | 40 | -0.171 | -0.167 | 0.073 | -0.137 | 0.110 |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| PIAS2 |   | 2 | 743 | 614 | 632 | 0.039 | -0.072 | 0.194 | 0.046 | -0.172 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| SRF |   | 1 | 1195 | 614 | 660 | -0.017 | -0.023 | 0.309 | -0.028 | 0.108 |
|---|
| NCBP2 |   | 3 | 557 | 614 | 623 | -0.087 | 0.170 | 0.311 | -0.055 | -0.052 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| FHL1 |   | 14 | 117 | 614 | 591 | -0.041 | 0.104 | 0.301 | undef | 0.266 |
|---|
| ITGB4 |   | 35 | 34 | 1 | 20 | 0.021 | 0.313 | -0.134 | 0.080 | -0.114 |
|---|
| HSPB1 |   | 22 | 69 | 1 | 25 | 0.053 | 0.169 | 0.017 | 0.179 | 0.161 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| CSNK1D |   | 6 | 301 | 614 | 602 | -0.222 | -0.044 | 0.170 | -0.088 | 0.013 |
|---|
| TUBB |   | 5 | 360 | 614 | 612 | -0.118 | 0.170 | 0.200 | undef | -0.045 |
|---|
GO Enrichment output for subnetwork 6634 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 2.638E-09 | 6.443E-06 |
|---|
| spindle organization | GO:0007051 |  | 9.603E-09 | 1.173E-05 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 1.087E-07 | 8.852E-05 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 2.543E-07 | 1.553E-04 |
|---|
| cell killing | GO:0001906 |  | 6.933E-07 | 3.387E-04 |
|---|
| regulation of nucleobase. nucleoside. nucleotide and nucleic acid transport | GO:0032239 |  | 1.106E-06 | 4.502E-04 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 2.415E-06 | 8.427E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 2.415E-06 | 7.374E-04 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 3.618E-06 | 9.822E-04 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 6.126E-06 | 1.496E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 1.303E-05 | 2.894E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 3.361E-09 | 8.086E-06 |
|---|
| spindle organization | GO:0007051 |  | 1.368E-08 | 1.646E-05 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 1.383E-07 | 1.11E-04 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 3.235E-07 | 1.946E-04 |
|---|
| cell killing | GO:0001906 |  | 8.814E-07 | 4.241E-04 |
|---|
| regulation of nucleobase. nucleoside. nucleotide and nucleic acid transport | GO:0032239 |  | 1.327E-06 | 5.321E-04 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 3.066E-06 | 1.054E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 3.066E-06 | 9.222E-04 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 4.593E-06 | 1.228E-03 |
|---|
| regulation of fat cell differentiation | GO:0045598 |  | 7.347E-06 | 1.768E-03 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 7.347E-06 | 1.607E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 1.123E-08 | 2.582E-05 |
|---|
| spindle organization | GO:0007051 |  | 5.284E-08 | 6.077E-05 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 2.769E-07 | 2.123E-04 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 7.173E-07 | 4.125E-04 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 1.968E-06 | 9.053E-04 |
|---|
| cell killing | GO:0001906 |  | 2.141E-06 | 8.207E-04 |
|---|
| regulation of nucleobase. nucleoside. nucleotide and nucleic acid transport | GO:0032239 |  | 3.282E-06 | 1.078E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 5.008E-06 | 1.44E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 6.405E-06 | 1.637E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 8.069E-06 | 1.856E-03 |
|---|
| regulation of fat cell differentiation | GO:0045598 |  | 1.137E-05 | 2.378E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| actin cytoskeleton reorganization | GO:0031532 |  | 2.255E-06 | 4.156E-03 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 7.33E-06 | 6.754E-03 |
|---|
| regulation of gene-specific transcription | GO:0032583 |  | 1.521E-05 | 9.346E-03 |
|---|
| response to insulin stimulus | GO:0032868 |  | 1.79E-05 | 8.247E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 2.429E-05 | 8.954E-03 |
|---|
| microtubule-based transport | GO:0010970 |  | 3.624E-05 | 0.01113278 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 3.624E-05 | 9.542E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 3.624E-05 | 8.35E-03 |
|---|
| positive regulation of specific transcription from RNA polymerase II promoter | GO:0010552 |  | 3.624E-05 | 7.422E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 3.682E-05 | 6.787E-03 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 5.354E-05 | 8.971E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| spindle assembly | GO:0051225 |  | 1.123E-08 | 2.582E-05 |
|---|
| spindle organization | GO:0007051 |  | 5.284E-08 | 6.077E-05 |
|---|
| natural killer cell mediated immunity | GO:0002228 |  | 2.769E-07 | 2.123E-04 |
|---|
| leukocyte mediated cytotoxicity | GO:0001909 |  | 7.173E-07 | 4.125E-04 |
|---|
| microtubule cytoskeleton organization | GO:0000226 |  | 1.968E-06 | 9.053E-04 |
|---|
| cell killing | GO:0001906 |  | 2.141E-06 | 8.207E-04 |
|---|
| regulation of nucleobase. nucleoside. nucleotide and nucleic acid transport | GO:0032239 |  | 3.282E-06 | 1.078E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 5.008E-06 | 1.44E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 6.405E-06 | 1.637E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 8.069E-06 | 1.856E-03 |
|---|
| regulation of fat cell differentiation | GO:0045598 |  | 1.137E-05 | 2.378E-03 |
|---|



