Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6622
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7534 | 3.542e-03 | 5.780e-04 | 7.718e-01 | 1.580e-06 |
|---|
| Loi | 0.2305 | 7.999e-02 | 1.128e-02 | 4.275e-01 | 3.859e-04 |
|---|
| Schmidt | 0.6740 | 0.000e+00 | 0.000e+00 | 4.426e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.466e-03 | 0.000e+00 |
|---|
| Wang | 0.2623 | 2.582e-03 | 4.294e-02 | 3.922e-01 | 4.348e-05 |
|---|
Expression data for subnetwork 6622 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
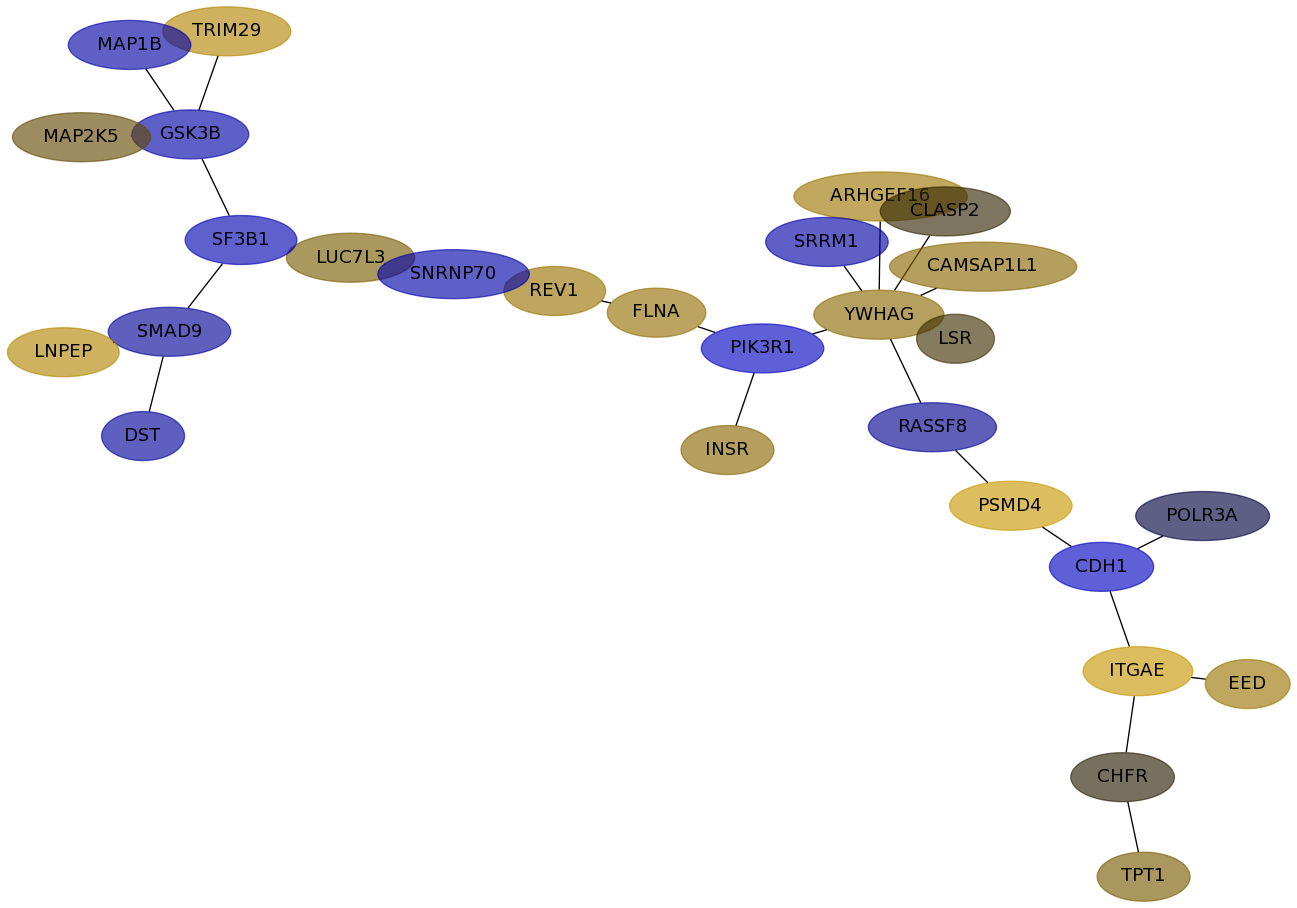
Score for each gene in subnetwork 6622 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| MAP2K5 |   | 2 | 743 | 296 | 336 | 0.024 | 0.039 | 0.130 | -0.036 | 0.096 |
|---|
| SMAD9 |   | 9 | 196 | 296 | 285 | -0.090 | -0.153 | 0.055 | 0.081 | -0.059 |
|---|
| SF3B1 |   | 18 | 86 | 296 | 280 | -0.154 | 0.126 | -0.073 | -0.131 | -0.204 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| LNPEP |   | 1 | 1195 | 296 | 366 | 0.162 | -0.234 | 0.099 | 0.107 | -0.003 |
|---|
| SRRM1 |   | 10 | 167 | 296 | 283 | -0.120 | 0.139 | 0.244 | 0.294 | -0.109 |
|---|
| CAMSAP1L1 |   | 4 | 440 | 296 | 321 | 0.062 | 0.083 | 0.113 | 0.151 | 0.150 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| PSMD4 |   | 11 | 148 | 296 | 282 | 0.285 | -0.243 | -0.043 | 0.087 | -0.045 |
|---|
| EED |   | 1 | 1195 | 296 | 366 | 0.092 | -0.074 | 0.067 | undef | -0.128 |
|---|
| POLR3A |   | 8 | 222 | 296 | 287 | -0.010 | -0.032 | undef | 0.091 | undef |
|---|
| YWHAG |   | 24 | 62 | 296 | 279 | 0.067 | 0.024 | undef | 0.266 | undef |
|---|
| ARHGEF16 |   | 10 | 167 | 296 | 283 | 0.096 | 0.036 | -0.047 | 0.200 | 0.038 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| CHFR |   | 1 | 1195 | 296 | 366 | 0.005 | 0.066 | -0.223 | 0.144 | -0.022 |
|---|
| RASSF8 |   | 5 | 360 | 296 | 310 | -0.078 | 0.121 | 0.174 | 0.114 | -0.092 |
|---|
| DST |   | 22 | 69 | 1 | 25 | -0.094 | 0.343 | 0.065 | -0.062 | 0.194 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| ITGAE |   | 1 | 1195 | 296 | 366 | 0.268 | -0.139 | 0.110 | 0.101 | 0.157 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| LUC7L3 |   | 16 | 104 | 296 | 281 | 0.043 | 0.237 | 0.175 | 0.135 | 0.074 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| CLASP2 |   | 1 | 1195 | 296 | 366 | 0.007 | 0.059 | 0.048 | 0.103 | -0.083 |
|---|
| SNRNP70 |   | 5 | 360 | 296 | 310 | -0.129 | 0.130 | -0.059 | -0.099 | 0.036 |
|---|
| LSR |   | 1 | 1195 | 296 | 366 | 0.010 | 0.058 | -0.043 | 0.060 | -0.052 |
|---|
| REV1 |   | 9 | 196 | 296 | 285 | 0.090 | -0.027 | 0.165 | 0.049 | 0.039 |
|---|
| TPT1 |   | 1 | 1195 | 296 | 366 | 0.042 | 0.149 | -0.046 | undef | -0.029 |
|---|
| TRIM29 |   | 8 | 222 | 296 | 287 | 0.161 | 0.240 | -0.150 | -0.077 | 0.024 |
|---|
GO Enrichment output for subnetwork 6622 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of microtubule polymerization or depolymerization | GO:0031110 |  | 2.227E-09 | 5.441E-06 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 8.794E-09 | 1.074E-05 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 2.232E-08 | 1.818E-05 |
|---|
| negative regulation of microtubule depolymerization | GO:0007026 |  | 5.822E-08 | 3.556E-05 |
|---|
| negative regulation of microtubule polymerization or depolymerization | GO:0031111 |  | 7.473E-08 | 3.651E-05 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 1.179E-07 | 4.801E-05 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 1.774E-07 | 6.191E-05 |
|---|
| positive regulation of transcription factor import into nucleus | GO:0042993 |  | 2.471E-06 | 7.547E-04 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 2.622E-06 | 7.118E-04 |
|---|
| negative regulation of protein complex disassembly | GO:0043242 |  | 3.169E-06 | 7.742E-04 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 3.796E-06 | 8.432E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 1.759E-07 | 4.232E-04 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 2.646E-07 | 3.183E-04 |
|---|
| regulation of microtubule polymerization or depolymerization | GO:0031110 |  | 2.646E-07 | 2.122E-04 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 8.476E-07 | 5.098E-04 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 1.847E-06 | 8.889E-04 |
|---|
| positive regulation of transcription factor import into nucleus | GO:0042993 |  | 3.342E-06 | 1.34E-03 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 3.896E-06 | 1.339E-03 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 5.637E-06 | 1.695E-03 |
|---|
| positive regulation of protein transport | GO:0051222 |  | 5.637E-06 | 1.507E-03 |
|---|
| negative regulation of microtubule depolymerization | GO:0007026 |  | 8.446E-06 | 2.032E-03 |
|---|
| positive regulation of protein import into nucleus | GO:0042307 |  | 8.446E-06 | 1.847E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 2.178E-07 | 5.01E-04 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 3.531E-07 | 4.06E-04 |
|---|
| regulation of microtubule polymerization or depolymerization | GO:0031110 |  | 3.531E-07 | 2.707E-04 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 1.339E-06 | 7.701E-04 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 2.432E-06 | 1.119E-03 |
|---|
| positive regulation of transcription factor import into nucleus | GO:0042993 |  | 3.677E-06 | 1.41E-03 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 4.591E-06 | 1.509E-03 |
|---|
| positive regulation of protein transport | GO:0051222 |  | 7.919E-06 | 2.277E-03 |
|---|
| positive regulation of protein import into nucleus | GO:0042307 |  | 8.704E-06 | 2.224E-03 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 9.653E-06 | 2.22E-03 |
|---|
| negative regulation of microtubule depolymerization | GO:0007026 |  | 1.105E-05 | 2.311E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 2.55E-08 | 4.7E-05 |
|---|
| positive regulation of transcription factor import into nucleus | GO:0042993 |  | 5.642E-07 | 5.199E-04 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 9.493E-07 | 5.832E-04 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 1.312E-06 | 6.043E-04 |
|---|
| positive regulation of protein transport | GO:0051222 |  | 1.768E-06 | 6.515E-04 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 2.332E-06 | 7.162E-04 |
|---|
| positive regulation of protein import into nucleus | GO:0042307 |  | 2.355E-06 | 6.2E-04 |
|---|
| regulation of microtubule polymerization or depolymerization | GO:0031110 |  | 6.129E-06 | 1.412E-03 |
|---|
| regulation of transcription factor import into nucleus | GO:0042990 |  | 1.26E-05 | 2.58E-03 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 1.547E-05 | 2.852E-03 |
|---|
| regulation of protein transport | GO:0051223 |  | 2.072E-05 | 3.472E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 2.178E-07 | 5.01E-04 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 3.531E-07 | 4.06E-04 |
|---|
| regulation of microtubule polymerization or depolymerization | GO:0031110 |  | 3.531E-07 | 2.707E-04 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 1.339E-06 | 7.701E-04 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 2.432E-06 | 1.119E-03 |
|---|
| positive regulation of transcription factor import into nucleus | GO:0042993 |  | 3.677E-06 | 1.41E-03 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 4.591E-06 | 1.509E-03 |
|---|
| positive regulation of protein transport | GO:0051222 |  | 7.919E-06 | 2.277E-03 |
|---|
| positive regulation of protein import into nucleus | GO:0042307 |  | 8.704E-06 | 2.224E-03 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 9.653E-06 | 2.22E-03 |
|---|
| negative regulation of microtubule depolymerization | GO:0007026 |  | 1.105E-05 | 2.311E-03 |
|---|



