Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6608
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7537 | 3.525e-03 | 5.740e-04 | 7.711e-01 | 1.560e-06 |
|---|
| Loi | 0.2305 | 7.996e-02 | 1.128e-02 | 4.275e-01 | 3.856e-04 |
|---|
| Schmidt | 0.6737 | 0.000e+00 | 0.000e+00 | 4.457e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.466e-03 | 0.000e+00 |
|---|
| Wang | 0.2622 | 2.594e-03 | 4.305e-02 | 3.927e-01 | 4.385e-05 |
|---|
Expression data for subnetwork 6608 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
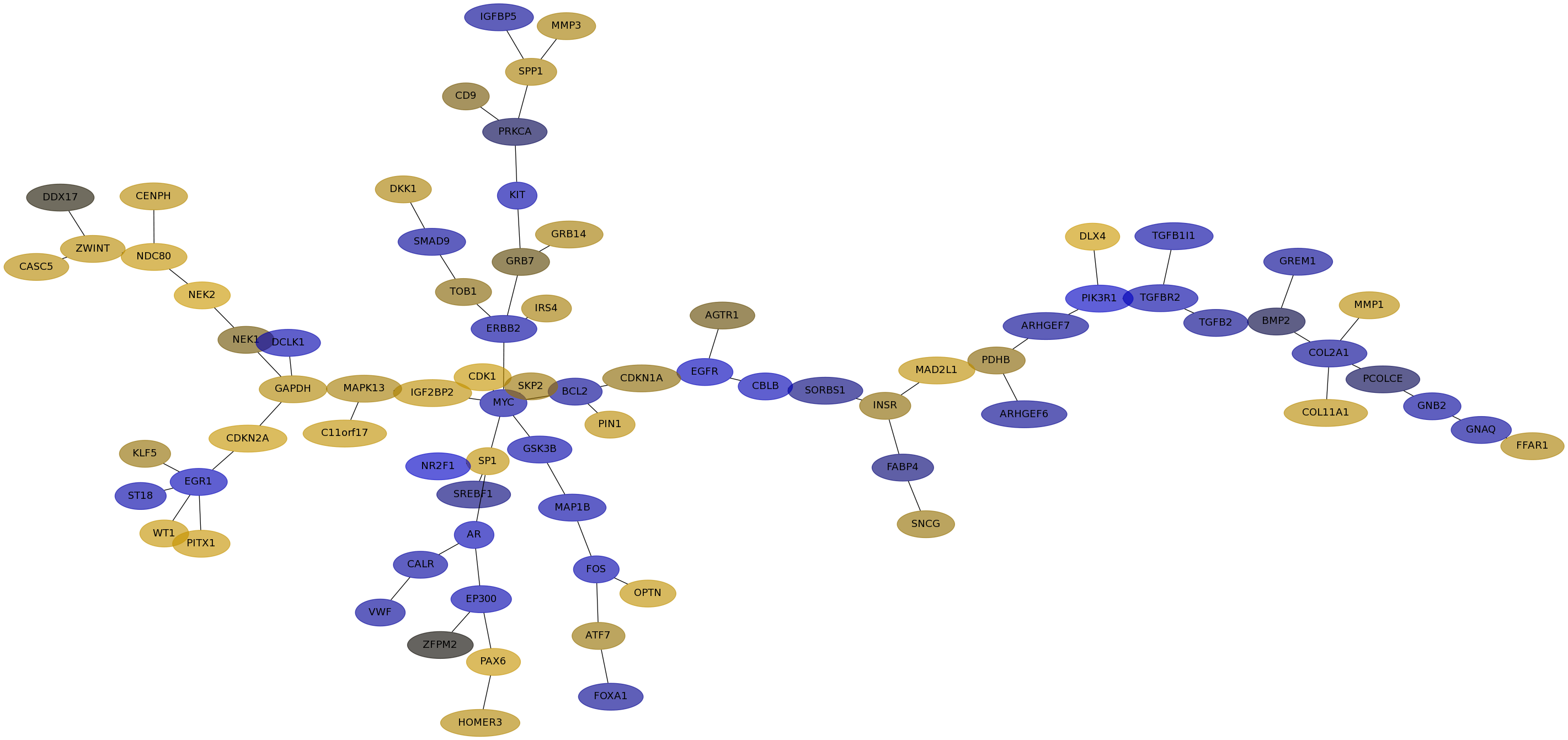
Score for each gene in subnetwork 6608 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| PAX6 |   | 22 | 69 | 318 | 312 | 0.259 | -0.003 | 0.112 | undef | 0.168 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| NDC80 |   | 2 | 743 | 525 | 547 | 0.246 | -0.229 | 0.060 | -0.005 | 0.044 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| COL11A1 |   | 3 | 557 | 525 | 540 | 0.191 | 0.149 | -0.023 | undef | 0.309 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| GNB2 |   | 11 | 148 | 525 | 505 | -0.091 | 0.082 | -0.062 | 0.208 | -0.062 |
|---|
| PIN1 |   | 24 | 62 | 179 | 181 | 0.194 | 0.040 | -0.040 | 0.065 | -0.048 |
|---|
| COL2A1 |   | 2 | 743 | 525 | 547 | -0.077 | -0.000 | 0.174 | -0.137 | 0.173 |
|---|
| ST18 |   | 16 | 104 | 236 | 227 | -0.131 | 0.028 | -0.088 | 0.186 | 0.151 |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| GAPDH |   | 11 | 148 | 141 | 143 | 0.159 | -0.040 | -0.053 | 0.088 | 0.029 |
|---|
| SMAD9 |   | 9 | 196 | 296 | 285 | -0.090 | -0.153 | 0.055 | 0.081 | -0.059 |
|---|
| CBLB |   | 2 | 743 | 525 | 547 | -0.168 | 0.266 | -0.248 | undef | 0.223 |
|---|
| MMP1 |   | 11 | 148 | 366 | 360 | 0.192 | 0.014 | 0.036 | 0.044 | 0.260 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| PDHB |   | 13 | 124 | 525 | 503 | 0.056 | 0.017 | 0.197 | -0.205 | 0.193 |
|---|
| OPTN |   | 7 | 256 | 1 | 47 | 0.226 | -0.099 | 0.015 | undef | 0.035 |
|---|
| DDX17 |   | 1 | 1195 | 525 | 574 | 0.004 | 0.048 | -0.062 | -0.141 | -0.187 |
|---|
| MMP3 |   | 6 | 301 | 179 | 205 | 0.121 | 0.171 | 0.081 | 0.198 | 0.192 |
|---|
| ARHGEF6 |   | 10 | 167 | 525 | 507 | -0.068 | -0.120 | 0.166 | undef | -0.115 |
|---|
| DCLK1 |   | 7 | 256 | 141 | 151 | -0.145 | -0.006 | -0.048 | 0.188 | -0.035 |
|---|
| CENPH |   | 1 | 1195 | 525 | 574 | 0.181 | -0.067 | undef | undef | undef |
|---|
| MAD2L1 |   | 8 | 222 | 236 | 236 | 0.208 | -0.130 | 0.116 | 0.032 | 0.063 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| MAPK13 |   | 1 | 1195 | 525 | 574 | 0.119 | -0.102 | 0.035 | -0.033 | 0.012 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| FABP4 |   | 6 | 301 | 236 | 239 | -0.036 | -0.028 | 0.227 | 0.072 | 0.107 |
|---|
| SNCG |   | 3 | 557 | 236 | 265 | 0.080 | -0.086 | 0.088 | -0.090 | -0.226 |
|---|
| PCOLCE |   | 2 | 743 | 525 | 547 | -0.016 | 0.150 | 0.032 | 0.004 | 0.127 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| GRB14 |   | 24 | 62 | 179 | 181 | 0.113 | -0.129 | 0.143 | -0.098 | 0.138 |
|---|
| ARHGEF7 |   | 25 | 59 | 83 | 92 | -0.074 | -0.061 | 0.208 | 0.028 | 0.015 |
|---|
| ATF7 |   | 28 | 47 | 179 | 175 | 0.085 | 0.115 | -0.088 | 0.172 | -0.092 |
|---|
| IGF2BP2 |   | 2 | 743 | 412 | 439 | 0.210 | -0.002 | 0.098 | -0.098 | -0.122 |
|---|
| TGFB2 |   | 7 | 256 | 525 | 515 | -0.063 | 0.244 | 0.226 | 0.079 | 0.015 |
|---|
| TGFBR2 |   | 7 | 256 | 525 | 515 | -0.115 | -0.028 | 0.136 | 0.105 | -0.190 |
|---|
| CALR |   | 4 | 440 | 525 | 520 | -0.105 | 0.002 | -0.061 | -0.094 | -0.075 |
|---|
| BMP2 |   | 11 | 148 | 525 | 505 | -0.011 | 0.097 | 0.148 | -0.114 | -0.069 |
|---|
| SPP1 |   | 26 | 52 | 83 | 91 | 0.130 | 0.119 | 0.184 | 0.204 | 0.153 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| SREBF1 |   | 9 | 196 | 478 | 460 | -0.042 | 0.232 | -0.195 | 0.140 | -0.074 |
|---|
| NEK2 |   | 8 | 222 | 141 | 149 | 0.292 | -0.106 | 0.020 | 0.083 | 0.165 |
|---|
| CD9 |   | 13 | 124 | 525 | 503 | 0.036 | -0.115 | -0.109 | 0.281 | -0.147 |
|---|
| WT1 |   | 33 | 37 | 1 | 21 | 0.254 | -0.039 | 0.036 | undef | 0.172 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| DLX4 |   | 10 | 167 | 236 | 234 | 0.291 | 0.057 | -0.004 | undef | -0.059 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| GREM1 |   | 9 | 196 | 525 | 508 | -0.073 | 0.200 | 0.028 | 0.225 | 0.261 |
|---|
| ZWINT |   | 1 | 1195 | 525 | 574 | 0.188 | -0.115 | 0.037 | -0.154 | -0.004 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| SKP2 |   | 29 | 45 | 1 | 23 | 0.092 | -0.043 | 0.237 | undef | -0.009 |
|---|
| ZFPM2 |   | 5 | 360 | 525 | 518 | 0.001 | 0.092 | 0.113 | -0.099 | 0.379 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| FFAR1 |   | 1 | 1195 | 525 | 574 | 0.135 | -0.008 | undef | undef | undef |
|---|
| IRS4 |   | 32 | 40 | 236 | 224 | 0.115 | -0.111 | -0.088 | 0.096 | 0.142 |
|---|
| NR2F1 |   | 25 | 59 | 83 | 92 | -0.252 | 0.113 | 0.133 | -0.116 | 0.207 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| DKK1 |   | 4 | 440 | 236 | 262 | 0.134 | 0.091 | -0.124 | 0.065 | 0.129 |
|---|
| HOMER3 |   | 2 | 743 | 525 | 547 | 0.160 | 0.124 | -0.083 | 0.072 | 0.094 |
|---|
| NEK1 |   | 15 | 111 | 141 | 139 | 0.032 | 0.184 | 0.049 | undef | 0.135 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| AGTR1 |   | 1 | 1195 | 525 | 574 | 0.025 | -0.058 | 0.174 | 0.058 | -0.007 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| PITX1 |   | 3 | 557 | 236 | 265 | 0.256 | 0.046 | 0.091 | -0.068 | 0.139 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| VWF |   | 5 | 360 | 525 | 518 | -0.091 | 0.072 | 0.152 | 0.127 | 0.100 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| GNAQ |   | 2 | 743 | 525 | 547 | -0.090 | 0.226 | 0.105 | undef | -0.062 |
|---|
| C11orf17 |   | 1 | 1195 | 525 | 574 | 0.232 | 0.023 | 0.056 | undef | -0.011 |
|---|
| CASC5 |   | 1 | 1195 | 525 | 574 | 0.177 | -0.245 | undef | undef | undef |
|---|
| TOB1 |   | 6 | 301 | 525 | 517 | 0.055 | 0.097 | -0.069 | 0.107 | -0.081 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
| FOXA1 |   | 27 | 50 | 179 | 176 | -0.072 | 0.071 | -0.015 | 0.214 | 0.005 |
|---|
GO Enrichment output for subnetwork 6608 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| urogenital system development | GO:0001655 |  | 3.21E-09 | 7.843E-06 |
|---|
| negative regulation of translation | GO:0017148 |  | 5.555E-08 | 6.786E-05 |
|---|
| positive regulation of inflammatory response | GO:0050729 |  | 7.26E-08 | 5.912E-05 |
|---|
| branching morphogenesis of a tube | GO:0048754 |  | 1.344E-07 | 8.207E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.497E-07 | 7.315E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.539E-07 | 6.265E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 2.297E-07 | 8.018E-05 |
|---|
| morphogenesis of a branching structure | GO:0001763 |  | 3.258E-07 | 9.949E-05 |
|---|
| cartilage development | GO:0051216 |  | 3.658E-07 | 9.929E-05 |
|---|
| regulation of cell cycle process | GO:0010564 |  | 5.153E-07 | 1.259E-04 |
|---|
| gland development | GO:0048732 |  | 5.557E-07 | 1.234E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| urogenital system development | GO:0001655 |  | 6.541E-09 | 1.574E-05 |
|---|
| negative regulation of translation | GO:0017148 |  | 8.782E-08 | 1.056E-04 |
|---|
| positive regulation of inflammatory response | GO:0050729 |  | 1.147E-07 | 9.201E-05 |
|---|
| branching morphogenesis of a tube | GO:0048754 |  | 2.305E-07 | 1.386E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.363E-07 | 1.137E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.638E-07 | 1.058E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 3.623E-07 | 1.245E-04 |
|---|
| morphogenesis of a branching structure | GO:0001763 |  | 5.571E-07 | 1.675E-04 |
|---|
| cartilage development | GO:0051216 |  | 5.571E-07 | 1.489E-04 |
|---|
| gland development | GO:0048732 |  | 8.152E-07 | 1.961E-04 |
|---|
| regulation of cell cycle process | GO:0010564 |  | 9.501E-07 | 2.078E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| urogenital system development | GO:0001655 |  | 1.498E-08 | 3.446E-05 |
|---|
| positive regulation of inflammatory response | GO:0050729 |  | 2.871E-07 | 3.302E-04 |
|---|
| branching morphogenesis of a tube | GO:0048754 |  | 6.06E-07 | 4.646E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 6.121E-07 | 3.52E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7E-07 | 3.22E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 9.551E-07 | 3.661E-04 |
|---|
| morphogenesis of a branching structure | GO:0001763 |  | 1.549E-06 | 5.088E-04 |
|---|
| cartilage development | GO:0051216 |  | 1.549E-06 | 4.452E-04 |
|---|
| gland development | GO:0048732 |  | 1.584E-06 | 4.048E-04 |
|---|
| regulation of cell cycle process | GO:0010564 |  | 2.235E-06 | 5.14E-04 |
|---|
| kidney development | GO:0001822 |  | 2.482E-06 | 5.189E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| gland development | GO:0048732 |  | 4.081E-08 | 7.521E-05 |
|---|
| urogenital system development | GO:0001655 |  | 2.211E-07 | 2.038E-04 |
|---|
| regulation of cell cycle process | GO:0010564 |  | 9.315E-07 | 5.723E-04 |
|---|
| negative regulation of translation | GO:0017148 |  | 1.078E-06 | 4.968E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.553E-06 | 5.724E-04 |
|---|
| brain development | GO:0007420 |  | 2.847E-06 | 8.746E-04 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 3.224E-06 | 8.488E-04 |
|---|
| prostate gland development | GO:0030850 |  | 3.362E-06 | 7.745E-04 |
|---|
| regulation of mitosis | GO:0007088 |  | 4.749E-06 | 9.724E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 4.749E-06 | 8.752E-04 |
|---|
| cell junction assembly | GO:0034329 |  | 5.05E-06 | 8.461E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| urogenital system development | GO:0001655 |  | 1.498E-08 | 3.446E-05 |
|---|
| positive regulation of inflammatory response | GO:0050729 |  | 2.871E-07 | 3.302E-04 |
|---|
| branching morphogenesis of a tube | GO:0048754 |  | 6.06E-07 | 4.646E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 6.121E-07 | 3.52E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7E-07 | 3.22E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 9.551E-07 | 3.661E-04 |
|---|
| morphogenesis of a branching structure | GO:0001763 |  | 1.549E-06 | 5.088E-04 |
|---|
| cartilage development | GO:0051216 |  | 1.549E-06 | 4.452E-04 |
|---|
| gland development | GO:0048732 |  | 1.584E-06 | 4.048E-04 |
|---|
| regulation of cell cycle process | GO:0010564 |  | 2.235E-06 | 5.14E-04 |
|---|
| kidney development | GO:0001822 |  | 2.482E-06 | 5.189E-04 |
|---|



