Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6607
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7538 | 3.521e-03 | 5.730e-04 | 7.709e-01 | 1.555e-06 |
|---|
| Loi | 0.2305 | 8.003e-02 | 1.130e-02 | 4.276e-01 | 3.866e-04 |
|---|
| Schmidt | 0.6737 | 0.000e+00 | 0.000e+00 | 4.456e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.463e-03 | 0.000e+00 |
|---|
| Wang | 0.2621 | 2.602e-03 | 4.312e-02 | 3.931e-01 | 4.410e-05 |
|---|
Expression data for subnetwork 6607 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
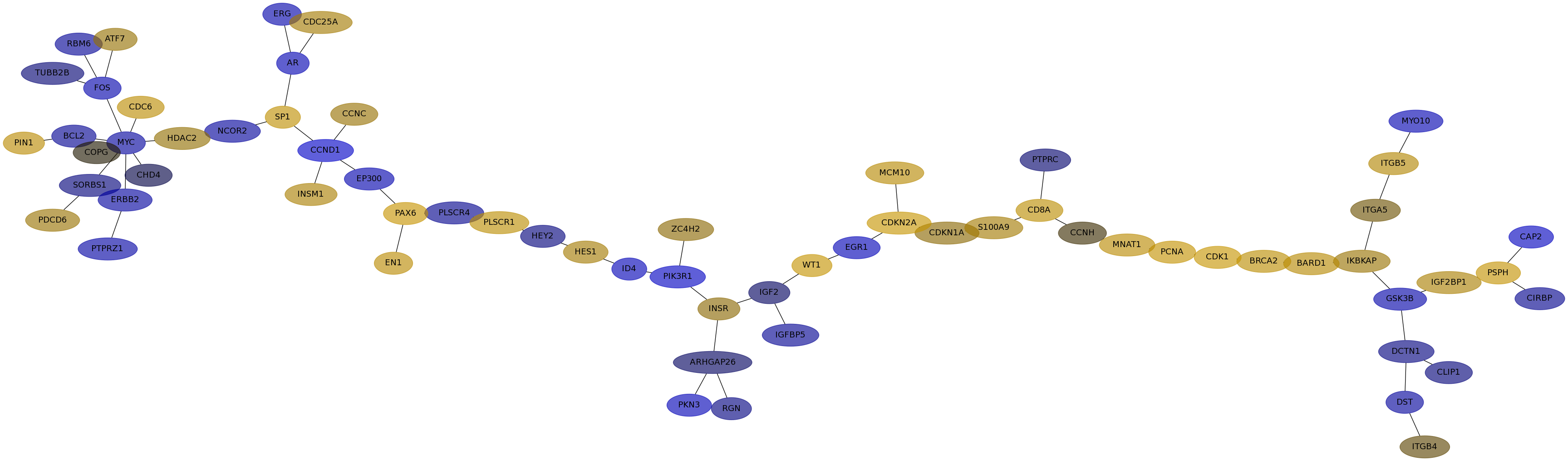
Score for each gene in subnetwork 6607 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| ARHGAP26 |   | 21 | 73 | 457 | 438 | -0.023 | 0.171 | -0.222 | 0.015 | -0.348 |
|---|
| CCNC |   | 5 | 360 | 570 | 562 | 0.085 | -0.053 | undef | 0.028 | undef |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| PAX6 |   | 22 | 69 | 318 | 312 | 0.259 | -0.003 | 0.112 | undef | 0.168 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| ID4 |   | 35 | 34 | 236 | 223 | -0.179 | 0.137 | 0.241 | undef | 0.036 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| RGN |   | 6 | 301 | 570 | 557 | -0.051 | -0.050 | 0.184 | 0.139 | -0.120 |
|---|
| DST |   | 22 | 69 | 1 | 25 | -0.094 | 0.343 | 0.065 | -0.062 | 0.194 |
|---|
| HEY2 |   | 8 | 222 | 570 | 555 | -0.045 | -0.021 | 0.398 | -0.024 | -0.058 |
|---|
| CCND1 |   | 26 | 52 | 1 | 24 | -0.258 | 0.074 | 0.117 | undef | 0.003 |
|---|
| PCNA |   | 15 | 111 | 1 | 32 | 0.243 | -0.158 | 0.027 | -0.099 | 0.003 |
|---|
| PIN1 |   | 24 | 62 | 179 | 181 | 0.194 | 0.040 | -0.040 | 0.065 | -0.048 |
|---|
| RBM6 |   | 2 | 743 | 570 | 593 | -0.082 | 0.265 | -0.004 | -0.096 | 0.111 |
|---|
| CHD4 |   | 2 | 743 | 318 | 346 | -0.012 | 0.079 | 0.092 | 0.062 | -0.070 |
|---|
| DCTN1 |   | 1 | 1195 | 570 | 625 | -0.050 | 0.223 | -0.055 | -0.048 | -0.120 |
|---|
| CCNH |   | 2 | 743 | 570 | 593 | 0.010 | -0.066 | 0.071 | -0.042 | 0.049 |
|---|
| CAP2 |   | 2 | 743 | 570 | 593 | -0.214 | -0.017 | 0.206 | 0.196 | 0.169 |
|---|
| CIRBP |   | 5 | 360 | 366 | 379 | -0.069 | 0.208 | 0.110 | 0.049 | -0.045 |
|---|
| CDC6 |   | 3 | 557 | 570 | 572 | 0.198 | -0.053 | 0.038 | -0.202 | 0.095 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| COPG |   | 5 | 360 | 570 | 562 | 0.004 | 0.080 | -0.158 | undef | 0.280 |
|---|
| PSPH |   | 20 | 81 | 366 | 356 | 0.240 | -0.019 | -0.102 | 0.358 | 0.025 |
|---|
| HDAC2 |   | 13 | 124 | 318 | 316 | 0.079 | -0.094 | 0.148 | 0.089 | 0.037 |
|---|
| NCOR2 |   | 32 | 40 | 318 | 290 | -0.096 | 0.211 | -0.163 | 0.158 | 0.114 |
|---|
| MCM10 |   | 5 | 360 | 179 | 208 | 0.178 | -0.245 | 0.015 | undef | -0.063 |
|---|
| PLSCR4 |   | 13 | 124 | 318 | 316 | -0.069 | 0.140 | 0.231 | 0.212 | 0.212 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| ATF7 |   | 28 | 47 | 179 | 175 | 0.085 | 0.115 | -0.088 | 0.172 | -0.092 |
|---|
| IKBKAP |   | 10 | 167 | 412 | 408 | 0.089 | 0.184 | 0.142 | 0.105 | -0.017 |
|---|
| ITGB5 |   | 4 | 440 | 412 | 419 | 0.163 | 0.208 | 0.081 | 0.278 | 0.148 |
|---|
| EN1 |   | 10 | 167 | 179 | 194 | 0.196 | 0.024 | 0.011 | -0.016 | 0.102 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| PTPRZ1 |   | 9 | 196 | 570 | 554 | -0.114 | 0.183 | 0.233 | -0.316 | -0.095 |
|---|
| WT1 |   | 33 | 37 | 1 | 21 | 0.254 | -0.039 | 0.036 | undef | 0.172 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| INSM1 |   | 23 | 66 | 570 | 545 | 0.135 | -0.184 | -0.188 | 0.237 | -0.032 |
|---|
| IGF2BP1 |   | 15 | 111 | 366 | 359 | 0.135 | -0.036 | undef | 0.190 | undef |
|---|
| S100A9 |   | 1 | 1195 | 570 | 625 | 0.117 | 0.043 | -0.110 | -0.020 | -0.045 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| CDC25A |   | 8 | 222 | 570 | 555 | 0.113 | -0.170 | 0.151 | -0.301 | -0.003 |
|---|
| HES1 |   | 4 | 440 | 570 | 565 | 0.115 | 0.157 | 0.010 | 0.214 | -0.106 |
|---|
| ERG |   | 9 | 196 | 179 | 195 | -0.134 | -0.026 | 0.235 | -0.027 | 0.182 |
|---|
| PLSCR1 |   | 10 | 167 | 412 | 408 | 0.202 | -0.159 | 0.091 | undef | -0.105 |
|---|
| MYO10 |   | 3 | 557 | 570 | 572 | -0.159 | 0.121 | -0.191 | 0.074 | 0.066 |
|---|
| ZC4H2 |   | 17 | 95 | 570 | 546 | 0.065 | 0.061 | 0.304 | -0.072 | 0.059 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| ITGA5 |   | 26 | 52 | 179 | 177 | 0.031 | 0.192 | 0.125 | 0.133 | 0.202 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| PTPRC |   | 9 | 196 | 83 | 99 | -0.031 | -0.204 | -0.050 | -0.179 | -0.068 |
|---|
| MNAT1 |   | 2 | 743 | 570 | 593 | 0.194 | 0.055 | 0.224 | 0.033 | 0.236 |
|---|
| CLIP1 |   | 1 | 1195 | 570 | 625 | -0.043 | 0.200 | -0.079 | -0.016 | 0.092 |
|---|
| CD8A |   | 1 | 1195 | 570 | 625 | 0.206 | -0.295 | -0.148 | -0.056 | -0.108 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| PDCD6 |   | 10 | 167 | 1 | 40 | 0.091 | -0.060 | undef | undef | undef |
|---|
| PKN3 |   | 18 | 86 | 179 | 187 | -0.181 | 0.009 | undef | 0.239 | undef |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| TUBB2B |   | 13 | 124 | 141 | 141 | -0.036 | 0.304 | 0.045 | 0.187 | 0.139 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| ITGB4 |   | 35 | 34 | 1 | 20 | 0.021 | 0.313 | -0.134 | 0.080 | -0.114 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
| IGF2 |   | 14 | 117 | 570 | 553 | -0.022 | 0.172 | 0.145 | 0.087 | 0.116 |
|---|
GO Enrichment output for subnetwork 6607 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.751E-08 | 4.277E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.311E-07 | 1.602E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 2.454E-07 | 1.999E-04 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 3.188E-07 | 1.947E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 7.162E-07 | 3.499E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.679E-07 | 3.127E-04 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 9.723E-07 | 3.393E-04 |
|---|
| phospholipid scrambling | GO:0017121 |  | 1.311E-06 | 4.002E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.458E-06 | 3.957E-04 |
|---|
| cell-substrate junction assembly | GO:0007044 |  | 1.924E-06 | 4.7E-04 |
|---|
| T cell differentiation | GO:0030217 |  | 2.43E-06 | 5.397E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.121E-08 | 5.102E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.549E-07 | 1.864E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 2.316E-07 | 1.857E-04 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 3.583E-07 | 2.155E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.494E-07 | 3.606E-04 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 8.604E-07 | 3.45E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 8.636E-07 | 2.968E-04 |
|---|
| interphase | GO:0051325 |  | 9.259E-07 | 2.785E-04 |
|---|
| L-serine biosynthetic process | GO:0006564 |  | 1.433E-06 | 3.83E-04 |
|---|
| phospholipid scrambling | GO:0017121 |  | 1.433E-06 | 3.447E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.464E-06 | 3.203E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.291E-08 | 9.87E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.739E-07 | 4.3E-04 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 3.748E-07 | 2.873E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 4.532E-07 | 2.606E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 1.52E-06 | 6.99E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.645E-06 | 6.306E-04 |
|---|
| interphase | GO:0051325 |  | 1.923E-06 | 6.317E-04 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 2.064E-06 | 5.933E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 2.814E-06 | 7.192E-04 |
|---|
| cell-substrate junction assembly | GO:0007044 |  | 4.097E-06 | 9.424E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 5.241E-06 | 1.096E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell-substrate junction assembly | GO:0007044 |  | 3.568E-07 | 6.575E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 2.263E-06 | 2.085E-03 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 2.574E-06 | 1.581E-03 |
|---|
| integrin-mediated signaling pathway | GO:0007229 |  | 3.452E-06 | 1.591E-03 |
|---|
| cell junction assembly | GO:0034329 |  | 3.907E-06 | 1.44E-03 |
|---|
| cell-matrix adhesion | GO:0007160 |  | 4.551E-06 | 1.398E-03 |
|---|
| focal adhesion formation | GO:0048041 |  | 5.518E-06 | 1.453E-03 |
|---|
| peptidyl-serine phosphorylation | GO:0018105 |  | 5.518E-06 | 1.271E-03 |
|---|
| cell-substrate adhesion | GO:0031589 |  | 7.561E-06 | 1.548E-03 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 9.635E-06 | 1.776E-03 |
|---|
| cell junction organization | GO:0034330 |  | 1.172E-05 | 1.963E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.291E-08 | 9.87E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.739E-07 | 4.3E-04 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 3.748E-07 | 2.873E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 4.532E-07 | 2.606E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 1.52E-06 | 6.99E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.645E-06 | 6.306E-04 |
|---|
| interphase | GO:0051325 |  | 1.923E-06 | 6.317E-04 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 2.064E-06 | 5.933E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 2.814E-06 | 7.192E-04 |
|---|
| cell-substrate junction assembly | GO:0007044 |  | 4.097E-06 | 9.424E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 5.241E-06 | 1.096E-03 |
|---|



