Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6604
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7538 | 3.520e-03 | 5.730e-04 | 7.709e-01 | 1.555e-06 |
|---|
| Loi | 0.2304 | 8.004e-02 | 1.130e-02 | 4.277e-01 | 3.868e-04 |
|---|
| Schmidt | 0.6737 | 0.000e+00 | 0.000e+00 | 4.458e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.465e-03 | 0.000e+00 |
|---|
| Wang | 0.2621 | 2.604e-03 | 4.314e-02 | 3.932e-01 | 4.417e-05 |
|---|
Expression data for subnetwork 6604 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
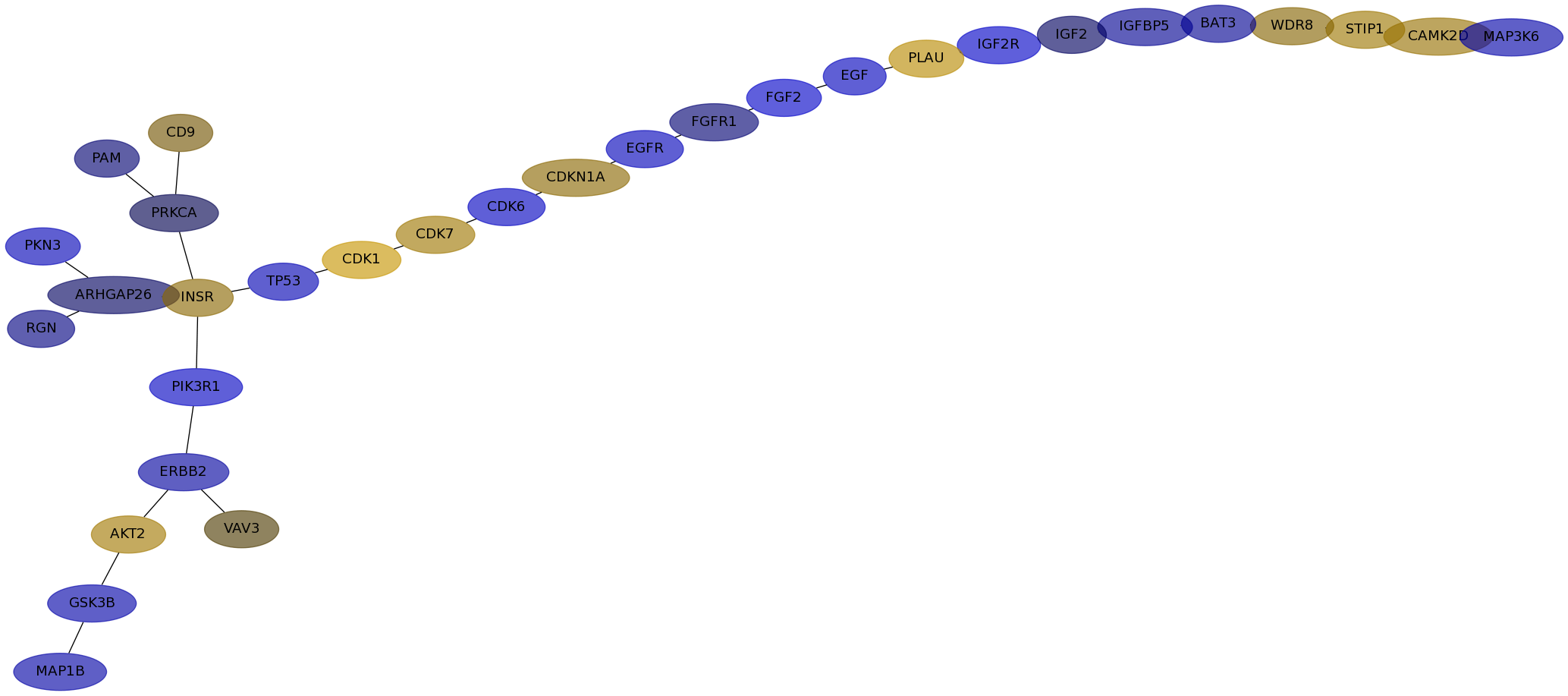
Score for each gene in subnetwork 6604 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| ARHGAP26 |   | 21 | 73 | 457 | 438 | -0.023 | 0.171 | -0.222 | 0.015 | -0.348 |
|---|
| VAV3 |   | 9 | 196 | 638 | 622 | 0.015 | 0.071 | -0.245 | 0.148 | 0.038 |
|---|
| IGF2R |   | 2 | 743 | 638 | 650 | -0.264 | -0.027 | -0.031 | 0.205 | 0.005 |
|---|
| STIP1 |   | 1 | 1195 | 638 | 686 | 0.102 | -0.038 | 0.002 | 0.183 | -0.123 |
|---|
| FGF2 |   | 1 | 1195 | 638 | 686 | -0.279 | 0.052 | 0.060 | -0.079 | -0.095 |
|---|
| AKT2 |   | 28 | 47 | 412 | 394 | 0.110 | -0.118 | -0.003 | 0.088 | -0.092 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| EGF |   | 12 | 140 | 638 | 620 | -0.212 | 0.182 | -0.276 | -0.060 | 0.005 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CDK7 |   | 7 | 256 | 318 | 328 | 0.105 | 0.184 | 0.017 | 0.078 | -0.097 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| MAP3K6 |   | 2 | 743 | 638 | 650 | -0.129 | 0.287 | -0.015 | 0.248 | 0.066 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| FGFR1 |   | 26 | 52 | 638 | 615 | -0.036 | 0.200 | 0.087 | undef | -0.015 |
|---|
| RGN |   | 6 | 301 | 570 | 557 | -0.051 | -0.050 | 0.184 | 0.139 | -0.120 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| CAMK2D |   | 2 | 743 | 638 | 650 | 0.085 | -0.149 | 0.000 | undef | undef |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| PLAU |   | 12 | 140 | 179 | 192 | 0.187 | 0.153 | 0.141 | 0.072 | 0.192 |
|---|
| PKN3 |   | 18 | 86 | 179 | 187 | -0.181 | 0.009 | undef | 0.239 | undef |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| WDR8 |   | 1 | 1195 | 638 | 686 | 0.057 | 0.004 | -0.040 | 0.192 | 0.034 |
|---|
| CD9 |   | 13 | 124 | 525 | 503 | 0.036 | -0.115 | -0.109 | 0.281 | -0.147 |
|---|
| PAM |   | 10 | 167 | 638 | 621 | -0.035 | 0.307 | 0.200 | 0.002 | 0.104 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| BAT3 |   | 4 | 440 | 236 | 262 | -0.078 | 0.072 | -0.057 | 0.110 | 0.069 |
|---|
| CDK6 |   | 16 | 104 | 318 | 314 | -0.229 | 0.107 | -0.028 | -0.118 | 0.104 |
|---|
| IGF2 |   | 14 | 117 | 570 | 553 | -0.022 | 0.172 | 0.145 | 0.087 | 0.116 |
|---|
GO Enrichment output for subnetwork 6604 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.153E-08 | 2.817E-05 |
|---|
| positive regulation of phosphorylation | GO:0042327 |  | 1.473E-07 | 1.8E-04 |
|---|
| positive regulation of phosphorus metabolic process | GO:0010562 |  | 1.705E-07 | 1.389E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 2.166E-07 | 1.323E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.166E-07 | 1.058E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 7.024E-07 | 2.86E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 7.024E-07 | 2.452E-04 |
|---|
| regulation of granule cell precursor proliferation | GO:0021936 |  | 8.444E-07 | 2.579E-04 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 8.444E-07 | 2.292E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 9.838E-07 | 2.404E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.152E-06 | 2.559E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 2.028E-08 | 4.88E-05 |
|---|
| positive regulation of phosphorylation | GO:0042327 |  | 2.063E-07 | 2.482E-04 |
|---|
| positive regulation of phosphorus metabolic process | GO:0010562 |  | 2.396E-07 | 1.922E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 2.59E-07 | 1.558E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.271E-07 | 1.574E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 6.785E-07 | 2.721E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 9.808E-07 | 3.371E-04 |
|---|
| regulation of granule cell precursor proliferation | GO:0021936 |  | 1.088E-06 | 3.273E-04 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 1.088E-06 | 2.91E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.373E-06 | 3.304E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.373E-06 | 3.003E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.763E-08 | 4.054E-05 |
|---|
| positive regulation of phosphorylation | GO:0042327 |  | 1.546E-07 | 1.778E-04 |
|---|
| positive regulation of phosphorus metabolic process | GO:0010562 |  | 1.832E-07 | 1.405E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 2.3E-07 | 1.322E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.621E-07 | 1.206E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 4.999E-07 | 1.916E-04 |
|---|
| ER overload response | GO:0006983 |  | 8.025E-07 | 2.637E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 9.362E-07 | 2.691E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.13E-06 | 2.889E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.353E-06 | 3.111E-04 |
|---|
| regulation of granule cell precursor proliferation | GO:0021936 |  | 1.401E-06 | 2.929E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 4.763E-09 | 8.778E-06 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 1.798E-08 | 1.657E-05 |
|---|
| regulation of MAP kinase activity | GO:0043405 |  | 2.997E-08 | 1.841E-05 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 6.297E-08 | 2.901E-05 |
|---|
| positive regulation of protein kinase activity | GO:0045860 |  | 1.494E-07 | 5.505E-05 |
|---|
| positive regulation of transferase activity | GO:0051347 |  | 1.715E-07 | 5.268E-05 |
|---|
| ER overload response | GO:0006983 |  | 4.874E-07 | 1.283E-04 |
|---|
| regulation of gene-specific transcription | GO:0032583 |  | 7.786E-07 | 1.794E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.253E-06 | 2.565E-04 |
|---|
| regulation of cell migration | GO:0030334 |  | 1.817E-06 | 3.348E-04 |
|---|
| regulation of locomotion | GO:0040012 |  | 3.096E-06 | 5.187E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.763E-08 | 4.054E-05 |
|---|
| positive regulation of phosphorylation | GO:0042327 |  | 1.546E-07 | 1.778E-04 |
|---|
| positive regulation of phosphorus metabolic process | GO:0010562 |  | 1.832E-07 | 1.405E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 2.3E-07 | 1.322E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.621E-07 | 1.206E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 4.999E-07 | 1.916E-04 |
|---|
| ER overload response | GO:0006983 |  | 8.025E-07 | 2.637E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 9.362E-07 | 2.691E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.13E-06 | 2.889E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.353E-06 | 3.111E-04 |
|---|
| regulation of granule cell precursor proliferation | GO:0021936 |  | 1.401E-06 | 2.929E-04 |
|---|



