Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6574
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7542 | 3.493e-03 | 5.670e-04 | 7.698e-01 | 1.525e-06 |
|---|
| Loi | 0.2306 | 7.986e-02 | 1.125e-02 | 4.272e-01 | 3.839e-04 |
|---|
| Schmidt | 0.6733 | 0.000e+00 | 0.000e+00 | 4.505e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.469e-03 | 0.000e+00 |
|---|
| Wang | 0.2619 | 2.640e-03 | 4.347e-02 | 3.947e-01 | 4.529e-05 |
|---|
Expression data for subnetwork 6574 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
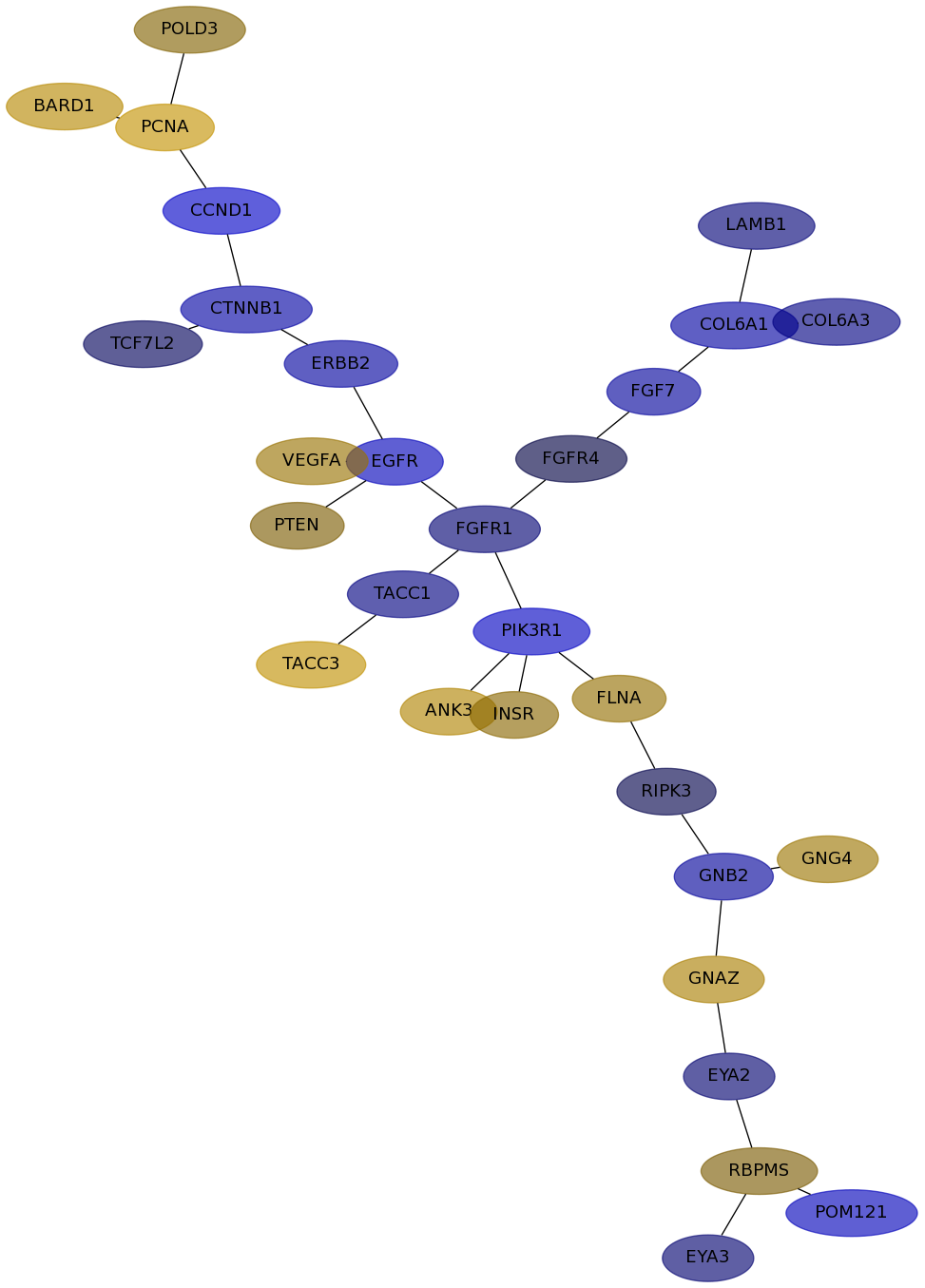
Score for each gene in subnetwork 6574 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| FGFR4 |   | 21 | 73 | 179 | 183 | -0.011 | 0.188 | -0.089 | -0.018 | -0.049 |
|---|
| TACC1 |   | 5 | 360 | 1077 | 1057 | -0.051 | 0.020 | -0.022 | 0.187 | 0.126 |
|---|
| RIPK3 |   | 19 | 85 | 179 | 186 | -0.014 | 0.040 | undef | 0.173 | undef |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| EYA3 |   | 9 | 196 | 318 | 324 | -0.034 | 0.053 | -0.020 | 0.188 | -0.142 |
|---|
| ANK3 |   | 6 | 301 | 83 | 105 | 0.153 | 0.034 | 0.001 | 0.094 | -0.038 |
|---|
| GNG4 |   | 5 | 360 | 1077 | 1057 | 0.095 | -0.139 | -0.169 | 0.129 | 0.106 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| POLD3 |   | 2 | 743 | 1077 | 1077 | 0.053 | -0.072 | 0.193 | 0.089 | 0.042 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| GNB2 |   | 11 | 148 | 525 | 505 | -0.091 | 0.082 | -0.062 | 0.208 | -0.062 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| TCF7L2 |   | 17 | 95 | 366 | 358 | -0.021 | 0.143 | 0.280 | undef | 0.101 |
|---|
| POM121 |   | 7 | 256 | 780 | 763 | -0.197 | 0.101 | 0.079 | undef | 0.017 |
|---|
| COL6A3 |   | 2 | 743 | 1077 | 1077 | -0.053 | 0.162 | 0.058 | 0.158 | 0.153 |
|---|
| FGFR1 |   | 26 | 52 | 638 | 615 | -0.036 | 0.200 | 0.087 | undef | -0.015 |
|---|
| CCND1 |   | 26 | 52 | 1 | 24 | -0.258 | 0.074 | 0.117 | undef | 0.003 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| PCNA |   | 15 | 111 | 1 | 32 | 0.243 | -0.158 | 0.027 | -0.099 | 0.003 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| PTEN |   | 27 | 50 | 141 | 135 | 0.045 | 0.055 | 0.017 | 0.183 | -0.041 |
|---|
| GNAZ |   | 15 | 111 | 478 | 457 | 0.134 | -0.116 | 0.308 | 0.267 | 0.011 |
|---|
| COL6A1 |   | 5 | 360 | 780 | 767 | -0.112 | 0.193 | -0.013 | undef | 0.039 |
|---|
| LAMB1 |   | 3 | 557 | 1077 | 1068 | -0.041 | 0.226 | -0.020 | -0.011 | 0.238 |
|---|
| FGF7 |   | 10 | 167 | 780 | 760 | -0.094 | 0.237 | 0.070 | 0.038 | 0.083 |
|---|
| RBPMS |   | 3 | 557 | 1010 | 1001 | 0.043 | 0.108 | 0.066 | 0.145 | -0.077 |
|---|
| EYA2 |   | 13 | 124 | 318 | 316 | -0.034 | 0.113 | -0.142 | undef | -0.019 |
|---|
| TACC3 |   | 4 | 440 | 1077 | 1065 | 0.232 | -0.144 | 0.014 | 0.046 | 0.004 |
|---|
GO Enrichment output for subnetwork 6574 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 9.766E-11 | 2.386E-07 |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 3.727E-10 | 4.552E-07 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 1.115E-09 | 9.084E-07 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 1.29E-09 | 7.876E-07 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 2.597E-09 | 1.269E-06 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 5.182E-09 | 2.11E-06 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 1.547E-08 | 5.4E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.955E-08 | 5.971E-06 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.406E-08 | 6.53E-06 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 2.426E-08 | 5.927E-06 |
|---|
| regulation of protein stability | GO:0031647 |  | 9.953E-08 | 2.21E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 5.303E-11 | 1.276E-07 |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 3.719E-10 | 4.474E-07 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 1.113E-09 | 8.928E-07 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 1.281E-09 | 7.704E-07 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 2.592E-09 | 1.247E-06 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 5.171E-09 | 2.074E-06 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 1.544E-08 | 5.308E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.93E-08 | 5.806E-06 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 2.421E-08 | 6.473E-06 |
|---|
| regulation of protein stability | GO:0031647 |  | 8.608E-08 | 2.071E-05 |
|---|
| endothelial cell migration | GO:0043542 |  | 1.722E-07 | 3.767E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 2.963E-08 | 6.816E-05 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 4.226E-07 | 4.86E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 5.341E-07 | 4.094E-04 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 8.431E-07 | 4.848E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.863E-07 | 4.537E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 9.863E-07 | 3.781E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.445E-06 | 4.747E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.472E-06 | 4.231E-04 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.472E-06 | 3.761E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.582E-06 | 3.639E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 2.349E-06 | 4.911E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cell migration | GO:0030334 |  | 4.68E-08 | 8.626E-05 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 5.631E-08 | 5.189E-05 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 6.297E-08 | 3.868E-05 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 8.55E-08 | 3.939E-05 |
|---|
| regulation of locomotion | GO:0040012 |  | 8.948E-08 | 3.298E-05 |
|---|
| regulation of cell motion | GO:0051270 |  | 8.948E-08 | 2.748E-05 |
|---|
| negative regulation of protein transport | GO:0051224 |  | 3.713E-07 | 9.776E-05 |
|---|
| maintenance of protein location in cell | GO:0032507 |  | 4.529E-07 | 1.043E-04 |
|---|
| maintenance of protein location | GO:0045185 |  | 6.553E-07 | 1.342E-04 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 7.786E-07 | 1.435E-04 |
|---|
| maintenance of location in cell | GO:0051651 |  | 9.184E-07 | 1.539E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 2.963E-08 | 6.816E-05 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 4.226E-07 | 4.86E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 5.341E-07 | 4.094E-04 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 8.431E-07 | 4.848E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.863E-07 | 4.537E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 9.863E-07 | 3.781E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.445E-06 | 4.747E-04 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.472E-06 | 4.231E-04 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.472E-06 | 3.761E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.582E-06 | 3.639E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 2.349E-06 | 4.911E-04 |
|---|



