Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6556
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7544 | 3.482e-03 | 5.650e-04 | 7.693e-01 | 1.513e-06 |
|---|
| Loi | 0.2307 | 7.965e-02 | 1.120e-02 | 4.267e-01 | 3.805e-04 |
|---|
| Schmidt | 0.6728 | 0.000e+00 | 0.000e+00 | 4.554e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.459e-03 | 0.000e+00 |
|---|
| Wang | 0.2617 | 2.660e-03 | 4.365e-02 | 3.956e-01 | 4.593e-05 |
|---|
Expression data for subnetwork 6556 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
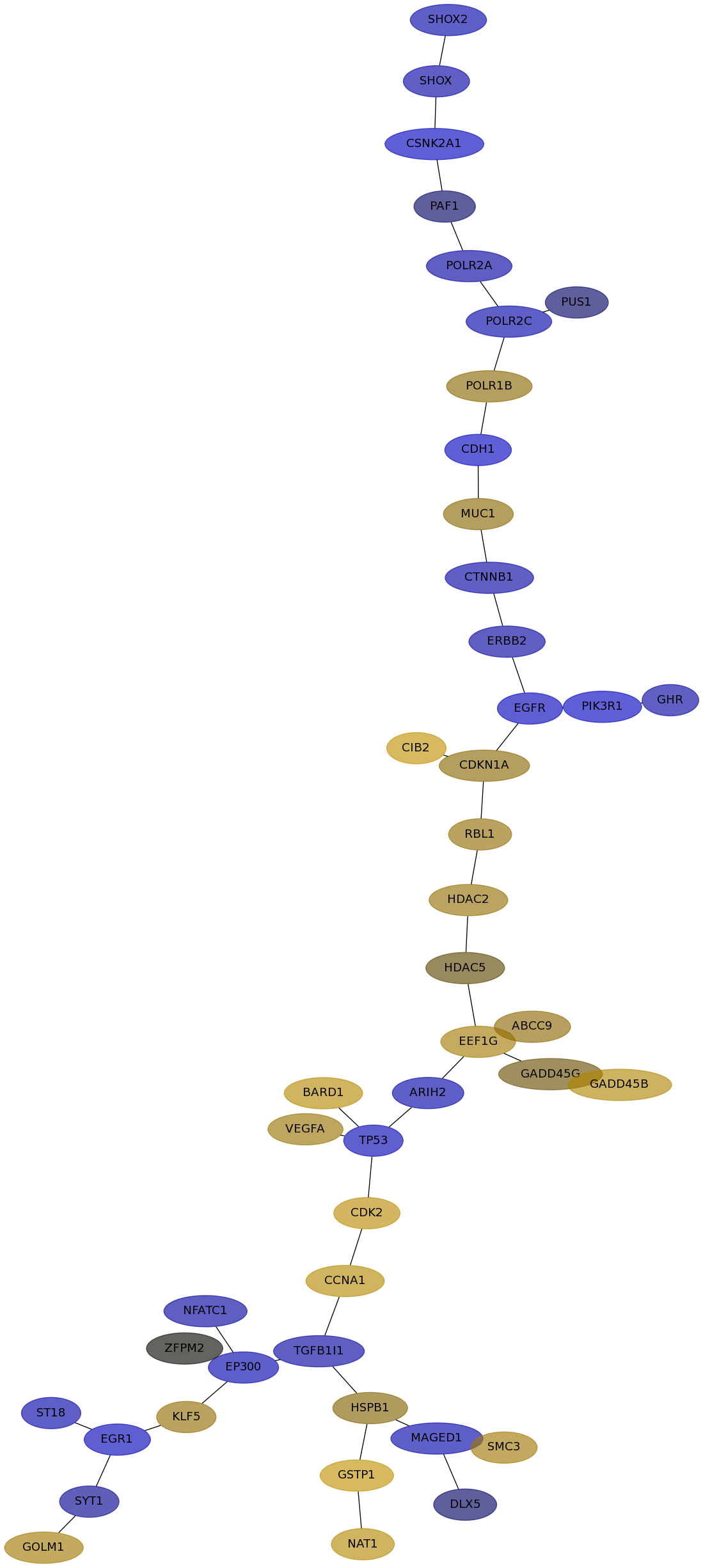
Score for each gene in subnetwork 6556 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| SHOX |   | 1 | 1195 | 1547 | 1563 | -0.113 | -0.061 | 0.000 | -0.004 | -0.092 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| MAGED1 |   | 5 | 360 | 842 | 828 | -0.141 | 0.284 | -0.152 | 0.240 | 0.045 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| CIB2 |   | 18 | 86 | 590 | 564 | 0.228 | -0.149 | 0.106 | 0.127 | 0.190 |
|---|
| CSNK2A1 |   | 3 | 557 | 1510 | 1486 | -0.215 | 0.029 | undef | 0.122 | undef |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| GSTP1 |   | 14 | 117 | 1 | 34 | 0.222 | -0.166 | 0.201 | 0.126 | -0.136 |
|---|
| EEF1G |   | 8 | 222 | 882 | 869 | 0.116 | -0.224 | 0.131 | 0.086 | 0.082 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| GOLM1 |   | 8 | 222 | 1534 | 1489 | 0.109 | 0.161 | -0.217 | 0.170 | -0.060 |
|---|
| ABCC9 |   | 1 | 1195 | 1547 | 1563 | 0.067 | -0.089 | 0.000 | 0.181 | -0.092 |
|---|
| SMC3 |   | 5 | 360 | 614 | 612 | 0.105 | -0.096 | 0.238 | 0.137 | 0.047 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| ST18 |   | 16 | 104 | 236 | 227 | -0.131 | 0.028 | -0.088 | 0.186 | 0.151 |
|---|
| POLR1B |   | 1 | 1195 | 1547 | 1563 | 0.062 | -0.036 | -0.079 | undef | 0.139 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| ZFPM2 |   | 5 | 360 | 525 | 518 | 0.001 | 0.092 | 0.113 | -0.099 | 0.379 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| SYT1 |   | 11 | 148 | 1 | 39 | -0.079 | 0.051 | 0.038 | 0.189 | -0.124 |
|---|
| NFATC1 |   | 1 | 1195 | 1547 | 1563 | -0.102 | -0.086 | 0.050 | 0.215 | 0.206 |
|---|
| HDAC5 |   | 2 | 743 | 882 | 892 | 0.022 | 0.029 | -0.165 | -0.125 | 0.009 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| ARIH2 |   | 5 | 360 | 236 | 242 | -0.131 | 0.196 | -0.071 | -0.127 | -0.016 |
|---|
| POLR2A |   | 6 | 301 | 1547 | 1510 | -0.107 | 0.247 | 0.091 | 0.040 | 0.141 |
|---|
| POLR2C |   | 1 | 1195 | 1547 | 1563 | -0.130 | 0.002 | -0.204 | undef | 0.168 |
|---|
| PAF1 |   | 1 | 1195 | 1547 | 1563 | -0.026 | 0.025 | -0.023 | 0.124 | -0.018 |
|---|
| MUC1 |   | 13 | 124 | 1547 | 1490 | 0.063 | 0.214 | -0.190 | undef | -0.132 |
|---|
| RBL1 |   | 21 | 73 | 1 | 27 | 0.078 | undef | undef | 0.051 | undef |
|---|
| NAT1 |   | 3 | 557 | 658 | 654 | 0.175 | 0.043 | 0.032 | -0.014 | 0.011 |
|---|
| GADD45B |   | 8 | 222 | 318 | 325 | 0.158 | 0.122 | 0.067 | 0.060 | 0.212 |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| DLX5 |   | 1 | 1195 | 1547 | 1563 | -0.026 | 0.044 | -0.151 | 0.094 | 0.140 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| PUS1 |   | 1 | 1195 | 1547 | 1563 | -0.026 | 0.045 | 0.044 | undef | 0.065 |
|---|
| CDK2 |   | 31 | 43 | 179 | 174 | 0.196 | -0.096 | 0.109 | 0.001 | 0.362 |
|---|
| SHOX2 |   | 1 | 1195 | 1547 | 1563 | -0.132 | 0.165 | 0.023 | 0.008 | 0.349 |
|---|
| HSPB1 |   | 22 | 69 | 1 | 25 | 0.053 | 0.169 | 0.017 | 0.179 | 0.161 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| HDAC2 |   | 13 | 124 | 318 | 316 | 0.079 | -0.094 | 0.148 | 0.089 | 0.037 |
|---|
GO Enrichment output for subnetwork 6556 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 4.801E-07 | 1.173E-03 |
|---|
| activation of MAPKKK activity | GO:0000185 |  | 7.831E-07 | 9.566E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.331E-06 | 1.084E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.182E-06 | 1.332E-03 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 3.264E-06 | 1.595E-03 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 6.38E-06 | 2.598E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 7.992E-06 | 2.789E-03 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 1.742E-05 | 5.319E-03 |
|---|
| activation of MAPKK activity | GO:0000186 |  | 2.59E-05 | 7.03E-03 |
|---|
| response to UV | GO:0009411 |  | 3.935E-05 | 9.613E-03 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 8.077E-05 | 0.01793734 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 5.111E-07 | 1.23E-03 |
|---|
| activation of MAPKKK activity | GO:0000185 |  | 9.406E-07 | 1.132E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.171E-06 | 9.393E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.62E-06 | 1.576E-03 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 3.919E-06 | 1.886E-03 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 5.584E-06 | 2.239E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 9.067E-06 | 3.117E-03 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 2.089E-05 | 6.283E-03 |
|---|
| activation of MAPKK activity | GO:0000186 |  | 2.564E-05 | 6.856E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 3.388E-05 | 8.153E-03 |
|---|
| response to UV | GO:0009411 |  | 4.297E-05 | 9.399E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 1.293E-06 | 2.974E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 2E-06 | 2.3E-03 |
|---|
| activation of MAPKKK activity | GO:0000185 |  | 2.274E-06 | 1.744E-03 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 6.323E-06 | 3.636E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 6.323E-06 | 2.909E-03 |
|---|
| activation of MAPKK activity | GO:0000186 |  | 4.023E-05 | 0.01542227 |
|---|
| cell cycle arrest | GO:0007050 |  | 6.524E-05 | 0.02143601 |
|---|
| response to UV | GO:0009411 |  | 7.609E-05 | 0.02187481 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.434E-04 | 0.03664296 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 1.434E-04 | 0.03297866 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 1.654E-04 | 0.03459118 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 2.367E-07 | 4.363E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 4.806E-07 | 4.429E-04 |
|---|
| positive regulation of protein kinase activity | GO:0045860 |  | 3.25E-06 | 1.997E-03 |
|---|
| positive regulation of transferase activity | GO:0051347 |  | 3.72E-06 | 1.714E-03 |
|---|
| activation of MAPKK activity | GO:0000186 |  | 1.771E-05 | 6.529E-03 |
|---|
| activation of protein kinase activity | GO:0032147 |  | 1.838E-05 | 5.647E-03 |
|---|
| lymphocyte differentiation | GO:0030098 |  | 3.619E-05 | 9.527E-03 |
|---|
| B cell differentiation | GO:0030183 |  | 8.555E-05 | 0.01970825 |
|---|
| leukocyte differentiation | GO:0002521 |  | 9.002E-05 | 0.01843326 |
|---|
| cell cycle arrest | GO:0007050 |  | 9.002E-05 | 0.01658994 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 9.002E-05 | 0.01508176 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 1.293E-06 | 2.974E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 2E-06 | 2.3E-03 |
|---|
| activation of MAPKKK activity | GO:0000185 |  | 2.274E-06 | 1.744E-03 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 6.323E-06 | 3.636E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 6.323E-06 | 2.909E-03 |
|---|
| activation of MAPKK activity | GO:0000186 |  | 4.023E-05 | 0.01542227 |
|---|
| cell cycle arrest | GO:0007050 |  | 6.524E-05 | 0.02143601 |
|---|
| response to UV | GO:0009411 |  | 7.609E-05 | 0.02187481 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.434E-04 | 0.03664296 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 1.434E-04 | 0.03297866 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 1.654E-04 | 0.03459118 |
|---|



